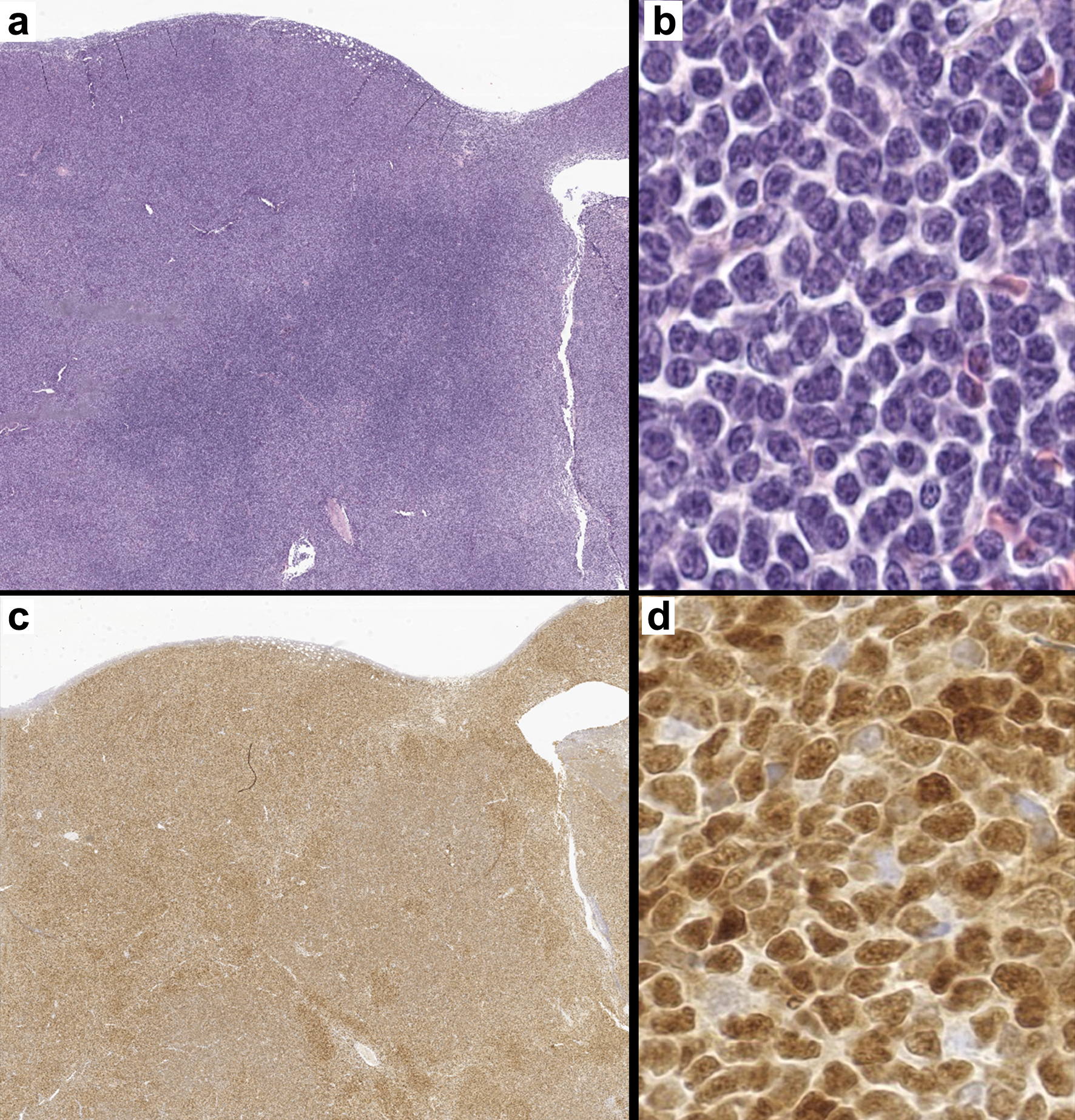
Figure 1. A representative case of mantle cell lymphoma. (a) Lymph node effaced by a neoplastic lymphoid process (H&E, × 7). (b) High-power image of a monotonous population of small to medium sized lymphocytes with irregular nuclear borders (H&E, × 400). (c) Cyclin D1 positivity confirming the diagnosis of mantle cell lymphoma (c: × 7, d: × 400). H&E: hematoxylin and eosin.
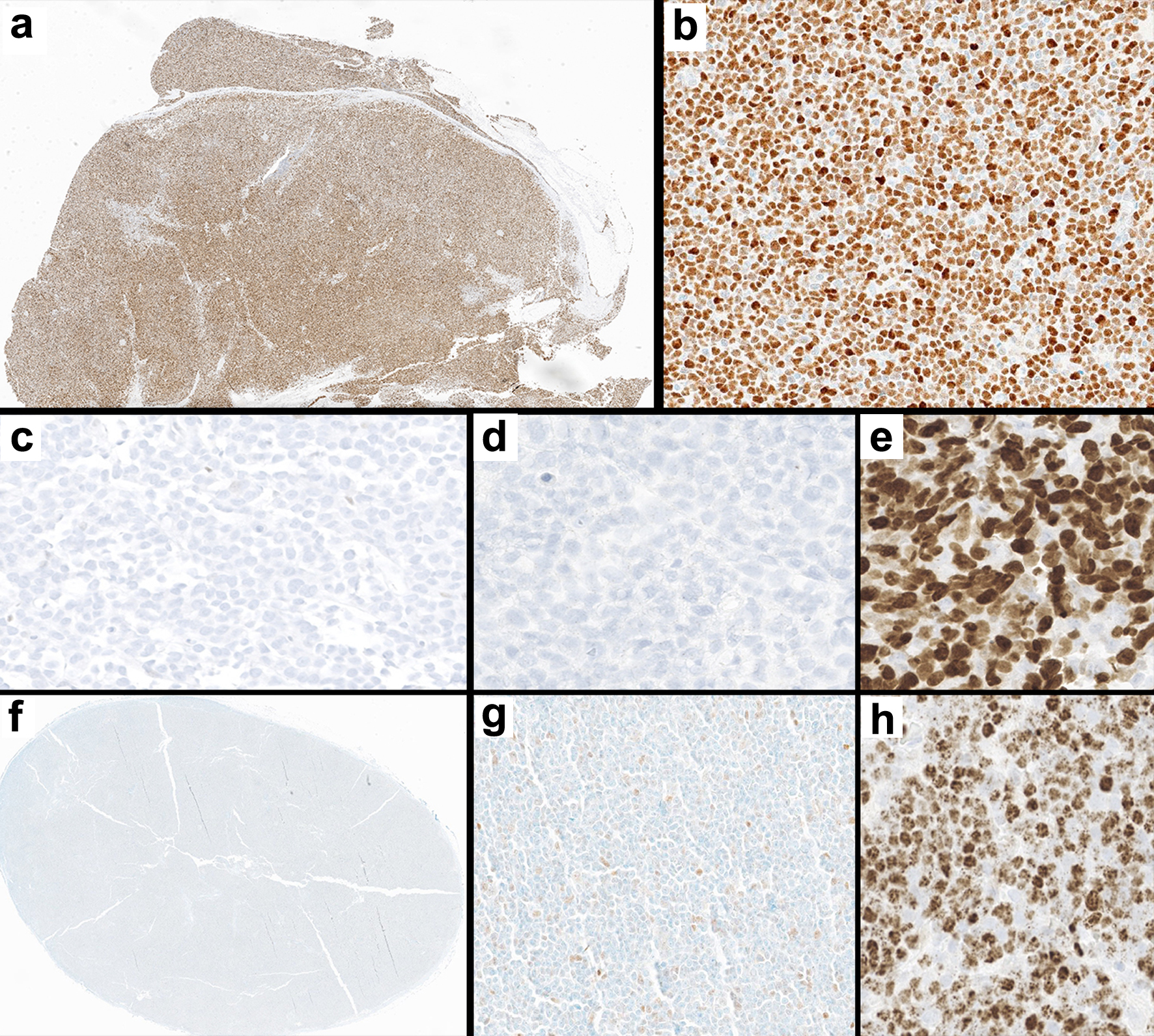
Figure 2. Immunohistochemical staining patterns in two TP53-pos cases (a-b and c-e) and a representative TP53-neg case (f-h). (a-b) Positive p53 IHC staining in TP53-pos MCL at low (a, × 10) and high (b, × 200) magnification. (c-e) Null p53 IHC in a TP53-pos MCL case (c, × 200), with rare non-neoplastic cells serving as internal control; this case was negative for SOX11 IHC (d, × 200) and had a high Ki-67 index (e, × 200). (f-h) Negative p53 IHC in TP53-neg MCL at low (f, × 10) and high (g, × 200) magnification; this tumor was positive for SOX11 IHC (h, × 200). IHC: immunohistochemistry; MCL: mantle cell lymphoma.

