Figures
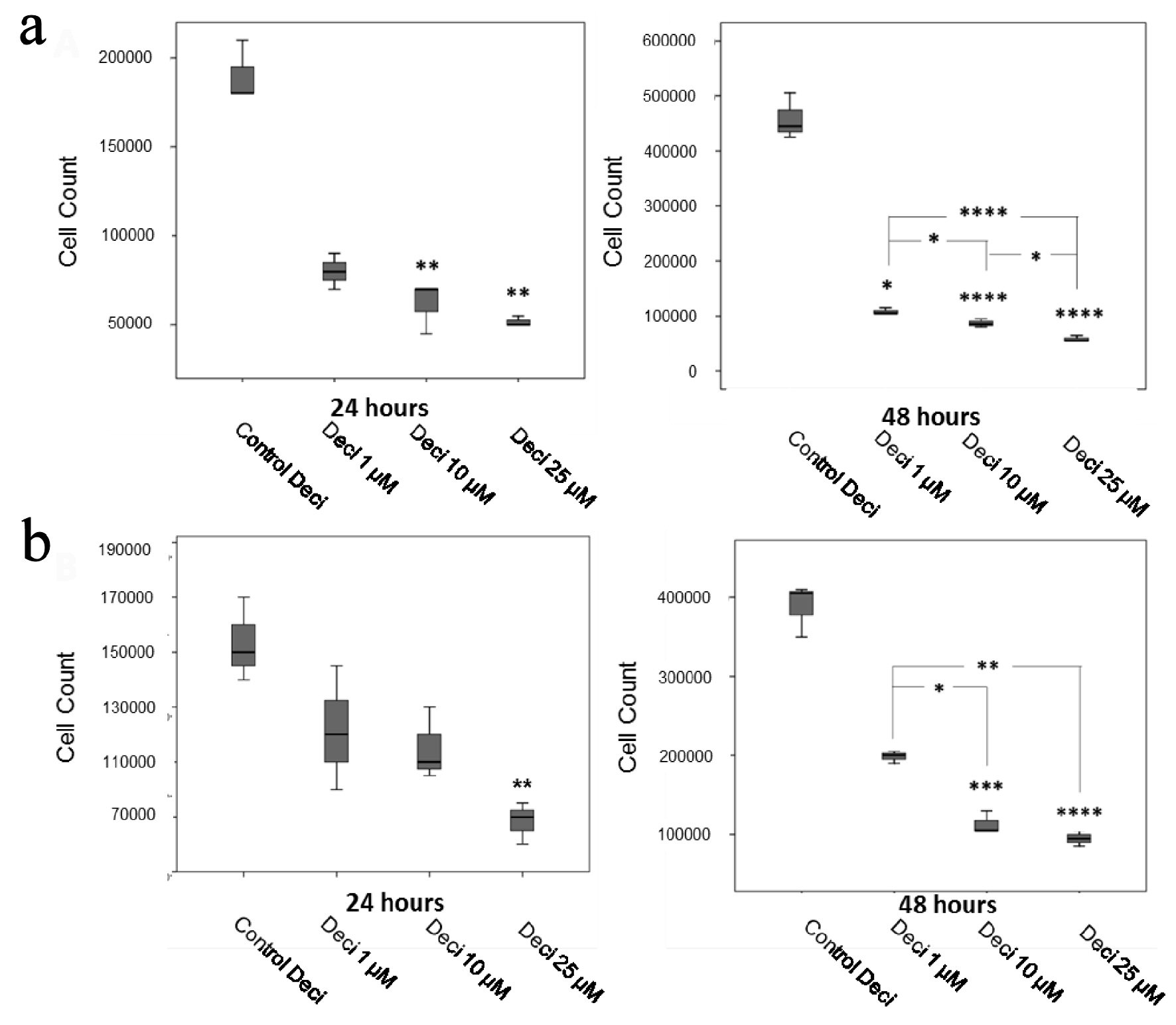
Figure 1. Decitabine dose curves in the KASUMI-1 cell line (a) and in the K-562 cell line (b). P values were determined using one-way ANOVA, followed by the Tukey post-hoc test (parametric), or using Kruskal-Wallis test, followed by Dunn’s multiple correlations test (non-parametric), using P < 0.05 for statistically significant differences. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Results are expressed as absolute cell counts.
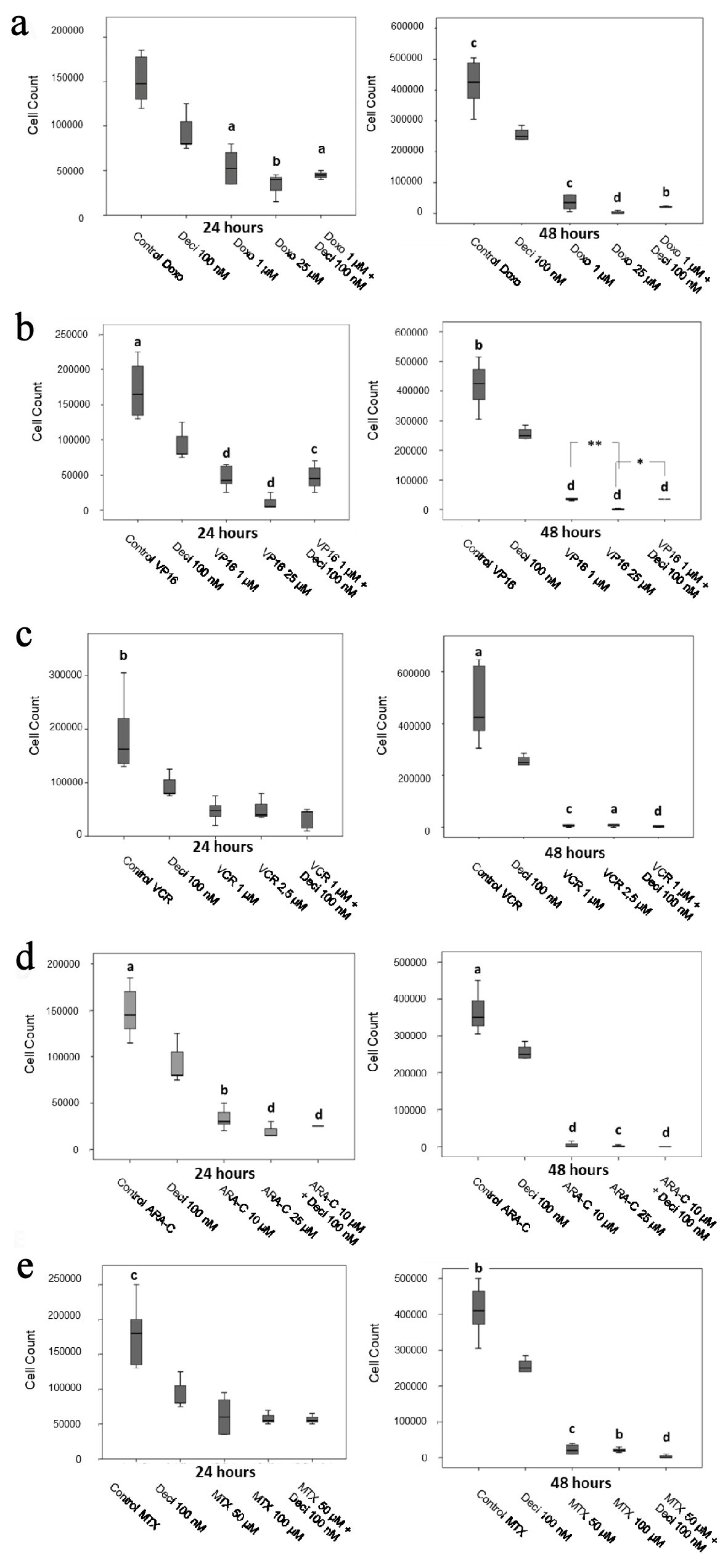
Figure 2. Cell counts after 24 and 48 h of treatment with doxorubicin (a), etoposide (b), vincristine (c), cytarabine (d) and methotrexate (e) in the KASUMI-1 cell line alone or in association with decitabine. The differences shown are in relation to treatment with decitabine. Values were determined using one-way ANOVA, followed by the Tukey post-hoc test (parametric), or Kruskal-Wallis test, followed by Dunn’s multiple correlations test (non-parametric), where P < 0.05 values determined statistically significant differences. a: P < 0.05, b: P < 0.01, c: P < 0.001, d: P < 0.0001. Results are expressed as absolute cell counts.
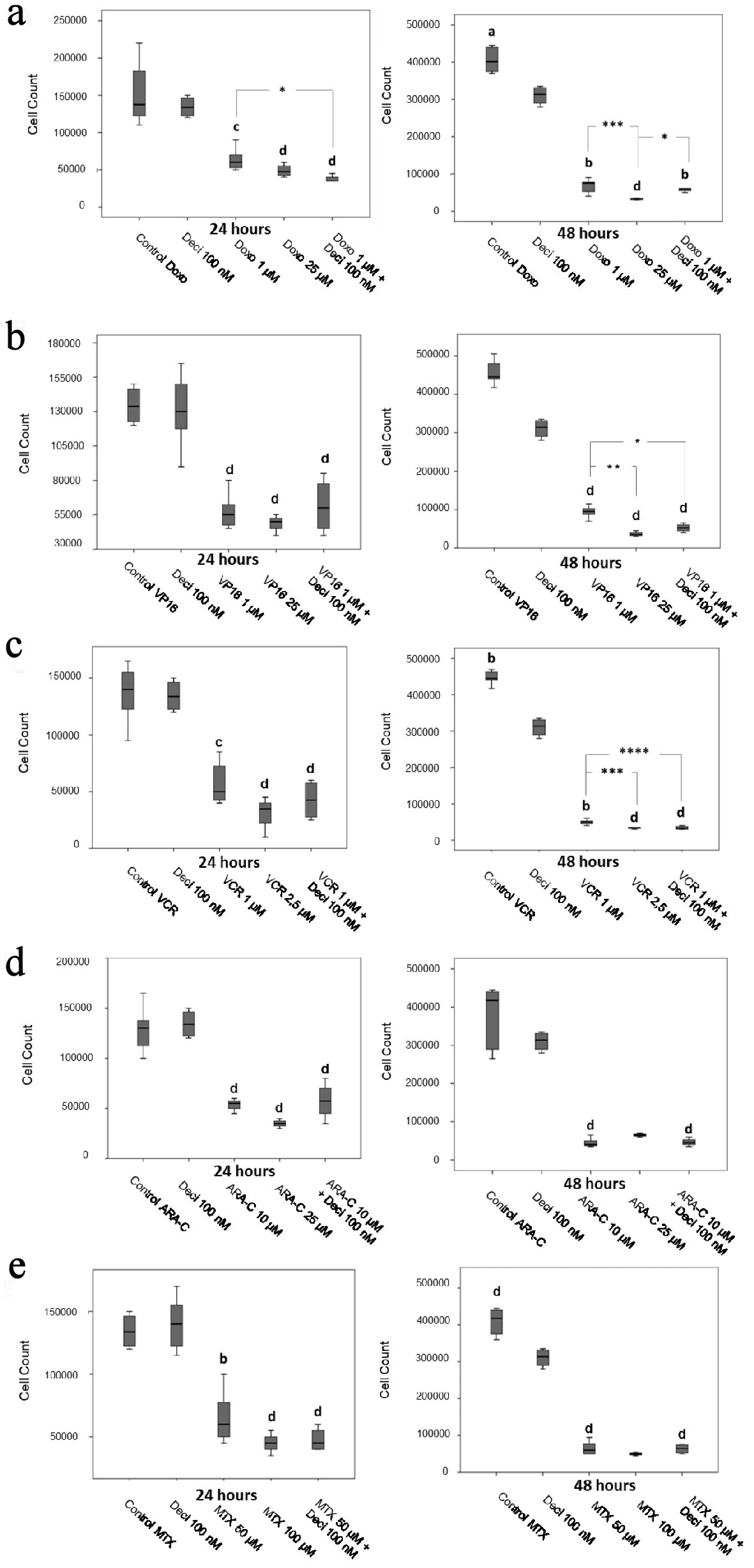
Figure 3. Cell counts after 24 and 48 h of treatment with doxorubicin (a), etoposide (b), vincristine (c), cytarabine (d) and methotrexate (e) in the K-562 cell line. The differences shown are in relation to treatment with decitabine. P values were determined using the Kruskal-Wallis test, followed by Dunn’s multiple correlation test, using P < 0.05 to determine statistically significant differences. */a: P < 0.05, b: P < 0.01, c/***: P < 0.001, d: P < 0.0001; values of P represented in letters are relative to decitabine; values of P represented in * are relative to the isolated chemotherapeutic treatment. Results are expressed as absolute cell counts.
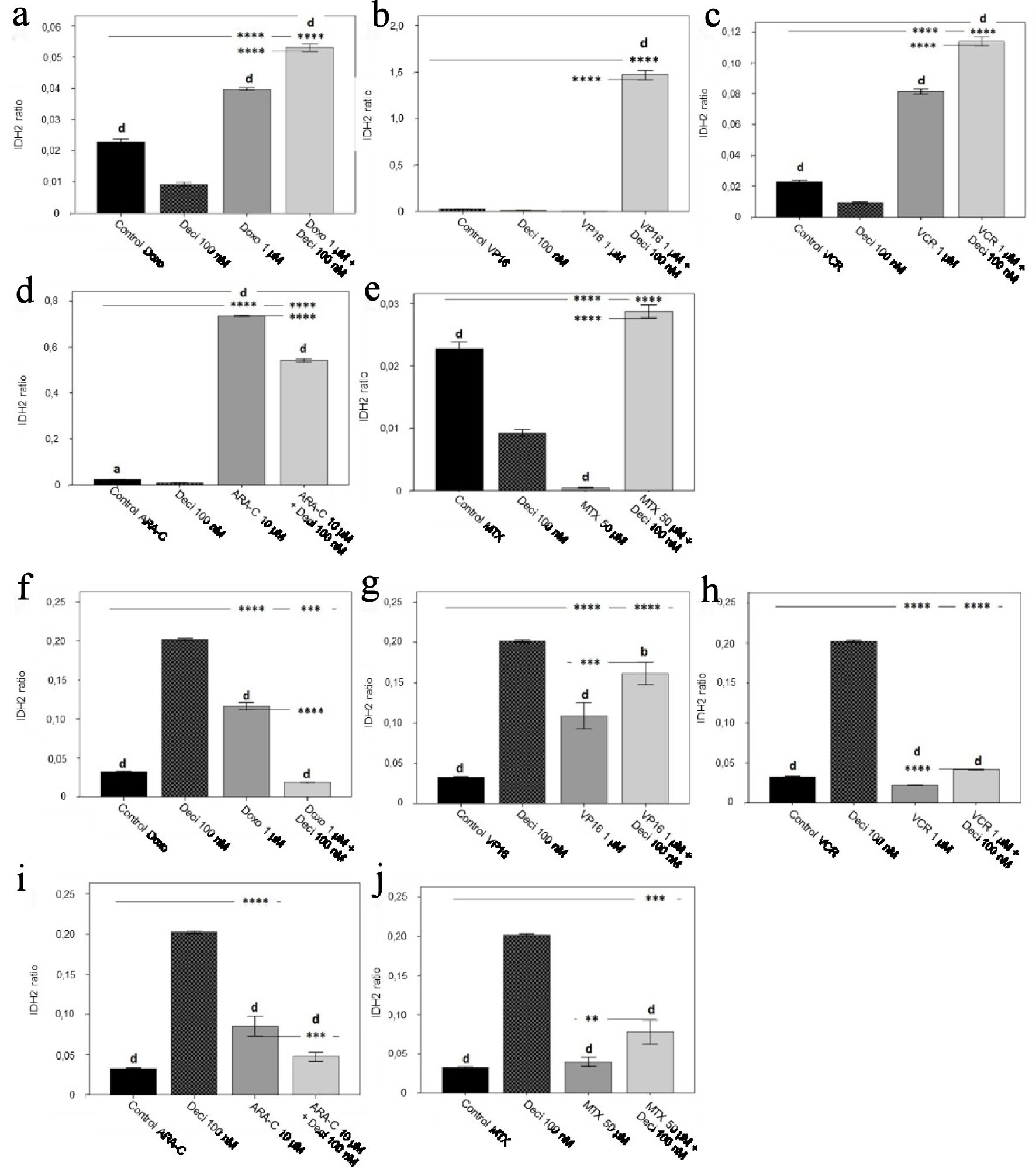
Figure 4. IDH2 gene expression levels in the KASUMI-1 cell line after treatment with (a) doxorubicin, (b) etoposide, (c) vincristine, (d) cytarabine, and (e) methotrexate, and in the K-562 cell line after treatment with (f) doxorubicin, (g) etoposide, (h) vincristine, (i) cytarabine, and (j) methotrexate. The differences shown are in relation to treatment with decitabine. P values were determined using one-way ANOVA, followed by the Tukey post-hoc test, using P < 0.05 to determine statistically significant differences. a/*: P < 0.05, b/**: P < 0.01, c/***: P < 0.001, d/****: P < 0.0001; values of P represented in letters are relative to decitabine; values of P represented in * are relative to the isolated control or chemotherapeutic agent.
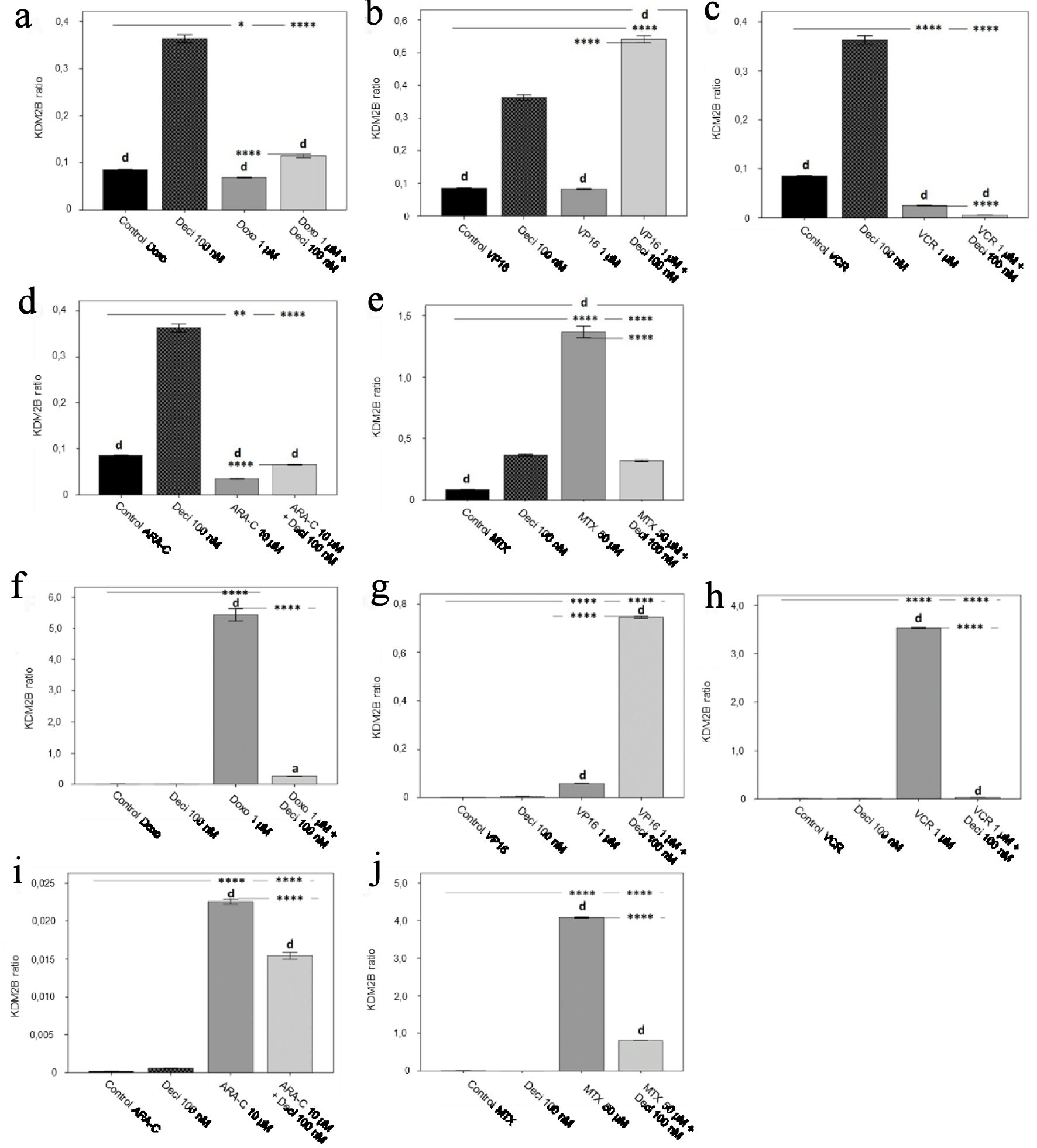
Figure 5. KDM2B gene expression levels in the KASUMI-1 cell line after treatment with (a) doxorubicin, (b) etoposide, (c) vincristine, (d) cytarabine, and (e) methotrexate, and in the K-562 cell line after treatment with (f) doxorubicin, (g) etoposide, (h) vincristine, (i) cytarabine, and (j) methotrexate. The differences shown are in relation to treatment with decitabine. P values were determined using one-way ANOVA, followed by the Tukey post-hoc test, using P < 0.05 to determine statistically significant differences. a/*: P < 0.05, b/**: P < 0.01, c/***: P < 0.001, d/****: P < 0.0001; values of P represented in letters are relative to decitabine; values of P represented in * are relative to the isolated control or chemotherapeutic agent.
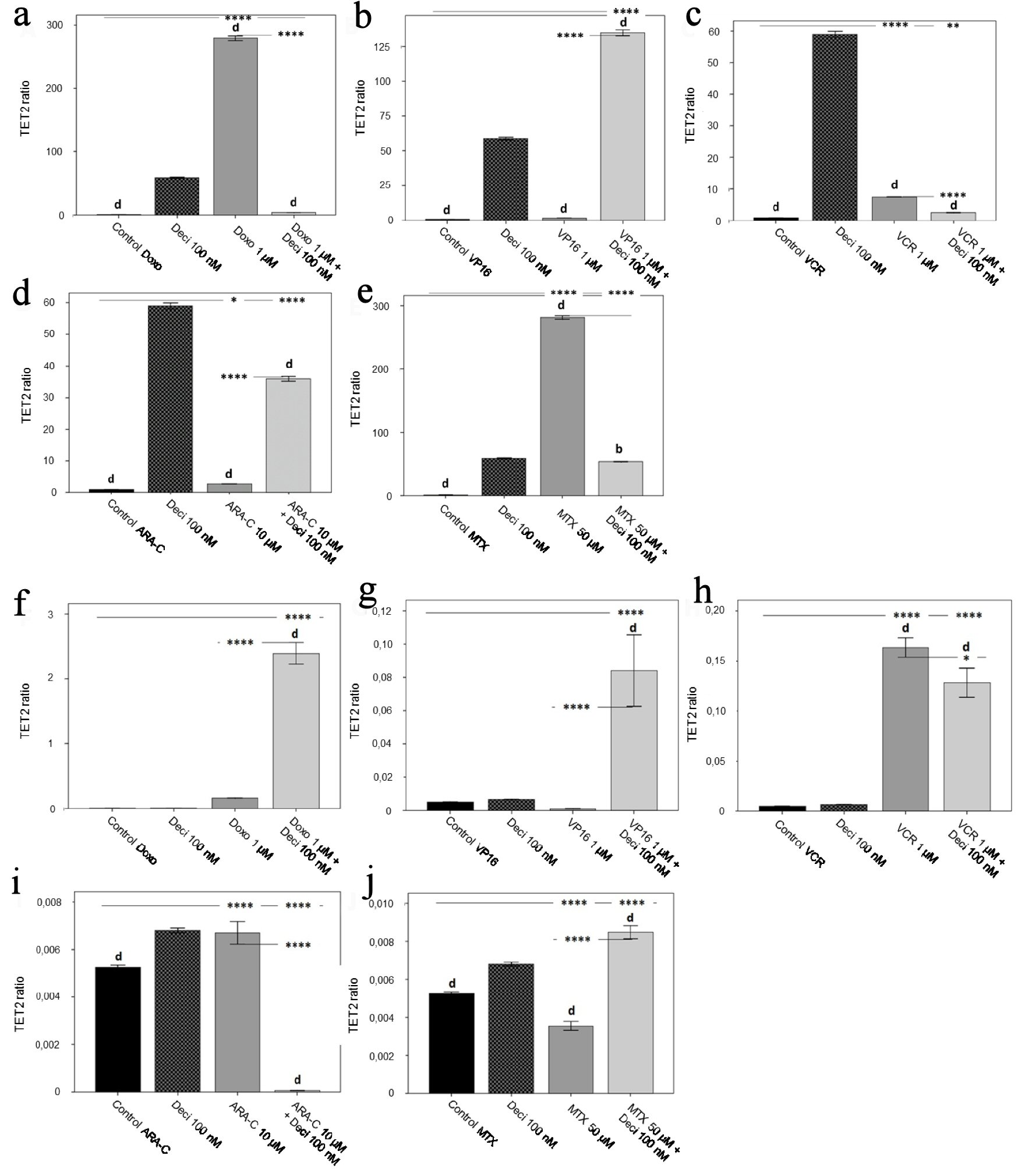
Figure 6. TET2 gene expression level in the KASUMI-1 cell line after treatment with (a) doxorubicin, (b) etoposide, (c) vincristine, (d) cytarabine, and (e) methotrexate, and in the K-562 cell line after treatment with (f) doxorubicin, (g) etoposide, (h) vincristine, (i) cytarabine, and (j) methotrexate. The differences shown are in relation to treatment with decitabine. P values were determined using one-way ANOVA, followed by the Tukey post-hoc test using P < 0.05 to determine statistically significant differences. a/*: P < 0.05, b/**: P < 0.01, c/***: P < 0.001, d/****: P < 0.0001; values of P represented in letters are relative to decitabine; values of P represented in * are relative to the isolated control or chemotherapeutic agent.
Tables
Table 1. Forward and Reverse Primers Used for Gene Amplification
| Gene | Primers 5’ - 3’ | Temperature (°C) | Size (bp) |
|---|
| IDH2 | Forward: ATGCCATCCAGAAGAAATGG | 59.89 | 175 |
| Reverse: TGAGCCACCATGTCATCAAT | 59.93 | |
| TET2 | Forward: TTGCAATGAGATACCCCACA | 59.92 | 200 |
| Reverse: TGCAAACCAACAAAGATGGA | 60.09 | |
| KDM2B | Forward: ACAACAAGGAAGGGCAGGAA | 59.44 | 196 |
| Reverse: CCAGGTTTGAGCCGCTTG | 59.04 | |
| ACTB | Forward: AAACTGGAACGGTGAAGGTG | 60.01 | 171 |
| Reverse: AGAGAAGTGGGGTGGCTTTT | 60.11 | |
| GAPDH | Forward: CTTTGTCAAGCTCATTTCCTGG | 54.20 | 133 |
| Reverse: TCTTCCTCTTGTGCTCTTGC | 54.90 | |
Table 2. Standardization of qPCR Reaction for IDH2, TET2 and KDM2B Genes in the K-562 Cell Line
| Reagent | IDH2 (60 °C) | TET2 (59 °C) | KDM2B (60 °C) |
|---|
| qPCR: quantitative real-time polymerase chain reaction; dNTPs: deoxyribonucleoside triphosphates; cDNA: complementary DNA. |
| Ultrapure water | 3.85 µL | 3.85 µL | 3.5 µL |
| Buffer (10 ×) | 2 µL | 2 µL | 2 µL |
| MgCl2 (50 nM) | 1.2 µL (3 nM) | 1.2 µL (3 nM) | 1.5 µL (3.75 nM) |
| dNTPs (10 mM) | - | - | 0.1 µL (0.05 mM) |
| dNTPs (5 mM) | 0.1 µL (0.025 mM) | 0.1 µL (0.025 mM) | - |
| Primer foward (10 mM) | 0.4 µL (0.2 mM) | 0.4 µL (0.2 mM) | 0.4 µL (0.2 mM) |
| Primer reverse (10 mM) | 0.4 µL (0.2 mM) | 0.4 µL (0.2 mM) | 0.4 µL (0.2 mM) |
| SYBR Green | 2 µL | 2 µL | 2 µL |
| Taq Platinum (5 U/µL) | 0.05 µL | 0.05 µL | 0.1 µL |
| cDNA (1:15) | 10 µL | 10 µL | 10 µL |
| Total | 20 µL | 20 µL | 20 µL |
Table 3. Standardization of qPCR Reaction for IDH2, TET2 and KDM2B Genes in the KASUMI-1 Cell Line
| Reagent | IDH2 (60 °C) | TET2 (59 °C) | KDM2B (60 °C) |
|---|
| qPCR: quantitative real-time polymerase chain reaction; dNTPs: deoxyribonucleoside triphosphates; cDNA: complementary DNA. |
| Ultrapure water | 3.6 µL | 3.6 µL | 3.5 µL |
| Buffer (10 ×) | 2 µL | 2 µL | 2 µL |
| MgCl2 (50 mM) | 1.2 µL (3 mM) | 1.2 µL (3 mM) | 1.5 µL (3.75 mM) |
| dNTPs (10 mM) | 0.1 µL (0.05 mM) | 0.1 µL (0.05 mM) | 0.1 µL (0.05 mM) |
| Primer forward (10 mM) | 0.5 µL (0.25 mM) | 0.5 µL (0.25 mM) | 0.4 µL (0.2 mM) |
| Primer reverse (10 mM) | 0.5 µL (0.25 mM) | 0.5 µL (0.25 mM) | 0.4 µL (0.2 mM) |
| SYBR Green | 2 µL | 2 µL | 2 µL |
| Taq Platinum (5 U/µL) | 0.1 µL | 0.1 µL | 0.1 µL |
| cDNA (1:15) | - | - | 10 µL |
| cDNA (1:45) | 10 µL | 10 µL | - |
| Total | 20 µL | 20 µL | 20 µL |





