| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website https://www.thejh.org |
Original Article
Volume 11, Number 3, June 2022, pages 87-91
Conventional Karyotyping and Fluorescence In Situ Hybridization for Detection of Chromosomal Abnormalities in Multiple Myeloma
Matthew Crabtreea , Jennifer Caia
, Xin Qinga, b
aDepartment of Pathology, Harbor-UCLA Medical Center, Torrance, CA 90502, USA
bCorresponding Author: Xin Qing, Hematology and Flow Cytometry Laboratories, Department of Pathology, Harbor-UCLA Medical Center, Torrance, CA 90502, USA
Manuscript submitted April 22, 2022, accepted June 18, 2022, published online June 27, 2022
Short title: Karyotyping and FISH in MM
doi: https://doi.org/10.14740/jh1007
| Abstract | ▴Top |
Background: Multiple myeloma (MM) is a genetically heterogeneous disease, with cytogenetic findings that determine disease behavior. Genetic abnormalities can be assessed by fluorescence in situ hybridization (FISH) analysis and/or G-banded karyotyping. The two methods produce unique and overlapping information, and the clinical utility of using both is investigated here.
Methods: Seventy patients diagnosed with MM at a hospital in Southern California were retrospectively reviewed for the FISH and G-banded karyotyping results obtained from bone marrow specimens.
Results: Karyotype was normal in 71% (50/70), abnormal in 27% (19/70), and inadequate in 1% (1/70). Among patients with abnormal karyotype, FISH provided additional information about genetic aberrations in 95% of cases (18/19). Among cases with abnormal FISH, karyotype provided additional information about genetic aberrations in 27% of cases (18/66). When numerical abnormalities were present (detected by FISH and/or karyotype), FISH detected them in 95% (54/57), of which karyotype missed 70% (38/54) of the time. Karyotyping detected numerical abnormalities in 33% (19/57), which FISH missed 16% (3/19) of the time.
Conclusions: Karyotyping and FISH analysis in MM each provide unique information. For most patients, performing both tests together will provide more information than either test alone.
Keywords: Multiple myeloma; Cytogenetics; FISH; Karyotype
| Introduction | ▴Top |
Multiple myeloma (MM) is a clonal bone marrow disease characterized by the neoplastic transformation of differentiated B cells. MM represents about 10-15% of all hematopoietic neoplasms and 1% of all cancer cases. The median age at diagnosis is 60 years old. Prognosis has been steadily improving over the last two decades alongside the introduction of new therapeutic strategies, which has prolonged the median survival from 3 to 8 - 10 years [1, 2, 3].
MM is diagnosed by bone marrow biopsy showing clonal bone marrow plasma cells ≥ 10% or biopsy showing plasmacytoma, and any one or more of the following myeloma-defining events: hypercalcemia, renal insufficiency, anemia, and bone lesions [1, 2].
Risk stratification in MM is a function of cytogenetic information and stratifies patients into three prognostic groups: high risk, intermediate risk, and standard risk [4-7]. This information can be acquired by conventional metaphase karyotyping or interphase fluorescence in situ hybridization (FISH) technology, and the two methodologies are partially overlapping in the information they provide of prognostic significance in MM [8]. We aimed to review a series of MM cases with both tests performed and summarize the extent to which these two tests provide unique information.
| Materials and Methods | ▴Top |
This research study was conducted retrospectively from data obtained for clinical purposes. Cytogenetic analysis was performed on 95 patients from 2013 to 2021 at Harbor-UCLA Medical Center in Southern California who had a bone marrow biopsy showing MM, monoclonal gammopathy of undetermined significance (MGUS), or residual disease after treatment. Of these 95, 70 satisfied the inclusion criteria of this study: clonal plasma cell percentage ≥ 10%, a diagnosis of MM for the first time, and both karyotyping and FISH performed on the corresponding specimen. Selected patients were aged 42 - 85 years (median 62) with 59% being male (41/70) and 41% being female (29/70) (Table 1). The 25 reviewed patients who were not included had a plasma cell percentage of < 10% (n = 17) or karyotyping was not performed (n = 20).
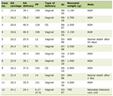 Click to view | Table 1. Demographics |
Karyotyping was performed at Quest Diagnostics Nichols Institute (San Juan Capistrano, CA). Fresh bone marrow aspirate samples were cultured as 48- and 72-h unstimulated cultures following standard cytogenetic methods. To increase the mitotic index of the cultures, stimulation with 2 µg/mL phorbol 12-myristate 13-acetate and 200 µL/mL phytohemagglutinin was also used. Chromosomal analysis was performed on cultured bone marrow samples using the standard G-banding technique. At least 20 metaphase cells were used for karyotyping.
FISH studies were performed at Quest Diagnostics Nichols Institute (San Juan Capistrano, CA). Two hundred to three hundred cells were counted and CD138 enrichment was performed whenever possible. Probes that were used included IGH (14q32), TP53 (17p13.1), D17Z1 (17p10), FGFR3 (4p16.3), CCND1 (11q13), MAF (16q23) (Abbott Molecular), MAFB (20q12) (Cytocell), DLEU (13q14.3), LAMP1 (13q34), CKS1B (1q21), CHD5 (1p36) (Kreatech), 9 (D9Z1), 11 (D11Z1), 15 (D15Z4) (SureFISH, Agilent DAKO, MetaSystems). Cutoff values were selected by Quest Diagnostics: +1q (4%), +9 (5%), +11 (6%), +15 (7%), 13q- (6%), -13 (4%), IGH rearrangement (8%), deletion TP53 (5%), t(4;14) (2%), t(11;14) (2%), t(14;16) (2%), and t(14;20) (2%).
This study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
| Results | ▴Top |
Among patients with MM, karyotype was normal in 71% (50/70), abnormal in 27% (19/70), and indeterminate due to inadequate specimen in 1% (1/70). FISH analysis detected abnormalities in 94% of cases (66/70). For patients with a normal karyotype (71%; 50/70), there was an abnormal FISH in 94% (47/50). For patients with normal FISH (6%; 4/70), there was an abnormal karyotype in 25% (1/4). Among patients with abnormal karyotype, FISH provided additional information about genetic aberrations in 95% of cases (18/19). Among cases with abnormal FISH, karyotype provided additional information about genetic aberrations in 27% of cases (18/66) (Table 2).
 Click to view | Table 2. Summary of Karyotype and FISH Findings |
Numerical abnormalities could be detected by either karyotyping or FISH and were seen in 81% (57/70). Among these, FISH detected the presence of one or more numerical abnormalities in 95% (54/57), of which karyotype missed 70% (38/54) of the time. Karyotyping detected numerical abnormalities in 33% (19/57) of overall cases with numerical abnormalities, for which FISH failed to identify any numerical abnormalities 16% (3/19) of the time (Table 3). Among these three cases, the numerical abnormalities missed by FISH were: +18 in one case, -Y in another, and +3, +11, -14, +15, -16, and +19 in the third.
 Click to view | Table 3. Frequency of Detection of Numerical and Structural Abnormalities |
In addition to numerical abnormalities, structural abnormalities were also detected by karyotyping and/or FISH, which included single arm gains or losses, translocations, or gene deletions. Structural abnormalities detected by either karyotyping or FISH were seen in 89% (62/70). Among these, FISH detected the presence of one or more structural abnormalities in 98% (61/62), of which karyotype missed 74% (45/61) of the time. The one case with a structural abnormality by karyotype (55,XY,+Y,t(2;8)(p12;q24.1),+3,+5,+7,+9,+11,+15,+19,+21[2]/46,XY[18]) and not by FISH (monosomy 13, gain 5, gain 9, gain 11, gain 15) was due to a t(2;8)(p12;q24.1) that only conventional karyotyping identified. Karyotyping detected structural abnormalities in 27% (17/62) of overall cases with structural abnormalities, and for which FISH missed 6% (1/17) of the time (Table 3).
The most common abnormalities of any kind affected chromosome 14 (76%; 53/70), with IGH rearrangements accounting for 47% (33/70) of all cases. Most common among them were t(11;14) (11%; 8/70), t(4;14) (9%; 6/70), and t(14;16) (1%; 1/70) (Table 4). Among the 20 cases with chromosome 14 abnormalities unrelated to IGH translocations, 90% (18/20) were monosomies or partial deletions of IGH, and 10% (2/20) were gains.
 Click to view | Table 4. Types of Abnormalities Detected by Karyotype or FISH and Their Frequencies |
The next most commonly aberrant chromosomes were chromosome 13 (56%; 39/70) and chromosome 11 (50%; 35/70). The most common abnormality of chromosome 13 was monosomy 13 (40%; 28/70), in chromosome 11 it was gain 11 (34%; 24/70), in chromosome 9, it was gain 9 (44%; 31/70), in chromosome 15, it was gain 15 (40%; 28/70), in chromosome 5, it was gain 5 (36%; 25/70), in chromosome 17, it was gain 17 (16%; 11/70), and in chromosome 4, it was t(4;14) (9%; 6/70) (Table 4).
| Discussion | ▴Top |
Conventional metaphase karyotyping is a well-established test in the clinical laboratory and is available all over the world. However, due to low proliferation rate of plasma cells and the resulted limited number of metaphases, abnormal karyotype is observed in only a subset of MM patients. In addition, karyotyping typically uses 400 band G-banding; each band represents approximately 10 Mbp and contains on the order of hundreds of genes, so karyotyping cannot detect small size genetic abnormalities. In contrast, interphase FISH studies are more sensitive and can reveal genetic aberrancies in most MM patients. In a study of 27 MM patients with G-banded karyotypes, 67% revealed additional genetic aberrations by the addition of FISH [9]. Our series had more success with FISH, with 94% (66/70) of MM patients revealing additional genetic abnormalities, including 95% (18/19) of those with an abnormal karyotype.
There are likely to be cytogenetic abnormalities in this specimen set that were not measured, given that selected FISH probes and their reporting are limited to targets of known clinical significance. Karyotyping can detect cytogenetic abnormalities at any location given that they are of sufficient size to be visible, and may identify potentially prognostically relevant chromosome abnormalities that are currently unknown in MM. Our data showed that karyotype detected additional genetic aberrations in 27% (19/70) of cases, including 27% (18/66) of those with abnormal FISH results.
In this data set, structural abnormalities (which are single arm gains or losses, translocations, or gene deletions) and numerical abnormalities (which are gains or losses of whole chromosomes) were each common. FISH was more sensitive (numerical: 95%; structural 98%) than karyotyping (numerical: 33%; structural 27%) in identifying cytogenetic abnormalities among cases that demonstrated their presence using either method. Karyotyping only identified one structural abnormality not identified by FISH, but did identify numerical abnormalities among 16% (3/19) of cases that did not show numerical abnormalities by FISH. Among these three cases, the numerical abnormalities missed were: +18 (case 1), -Y (case 2), and +3, +11, -14, +15, -16, +19 (case 3). Each of these changes are documented as common genetic lesions in MM [10], but less is known about the independent prognostic significance of each. Monosomy 16 and loss of Y are associated with reduced overall survival [9]. Trisomies of 3, 7, 9, 11, 15, 19 and/or 21 (four of which were missed here) have better response rates to treatment and longer survival than patients with other aneuploidies [11]. Of the 53 patients for whom revised International Staging System (R-ISS) could be calculated, 13 had high-risk cytogenetics and FISH alone would have been sufficient to account for their high-risk status in every case. While R-ISS is a function of del(17p), t(4;14), and t(14;16), the individual role of each observed chromosomal abnormality should be evaluated in future studies to determine their relationship to disease pathogenesis and clinical course, and this in turn should inform laboratories on the suitable applications of conventional karyotyping and the development of more prognostically informative FISH panels.
Conclusion
This study compares the results of karyotyping and FISH analysis for patients with MM. The results show that most MM patients with normal karyotype have demonstrable cytogenetic abnormalities with FISH, and routine FISH analysis appears to be an efficient method for detection of prognostically relevant chromosomal abnormalities in MM. Conversely, FISH alone without karyotyping may occasionally miss cytogenetic abnormalities of prognostic significance (such as monosomy 16, loss of Y, or various trisomies, as examples observed in this dataset). Therefore, performing both tests together will add valuable information in the cytogenetic workup of MM, and may detect new cytogenetic aberrancies that have potential prognostic significance.
Acknowledgments
None to declare.
Financial Disclosure
None to declare.
Conflict of Interest
None to declare.
Informed Consent
Not applicable.
Author Contributions
Matthew Crabtree aggregated data, analyzed data, reported findings, and wrote the manuscript. Jennifer Cai aggregated data and reviewed the manuscript. Xin Qing oversaw all histopathologic work, analyzed data, and reviewed the manuscript.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
| References | ▴Top |
- Hamdaoui H, Benlarroubia O, Ait Boujmia OK, Mossafa H, Ouldim K, Belkhayat A, Smyej I, et al. Cytogenetic and FISH analysis of 93 multiple myeloma Moroccan patients. Mol Genet Genomic Med. 2020;8(9):e1363.
doi pubmed - Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. WHO Classification of Tumours, revised 4th Edition. Volume 2.
- Gulla A, Anderson KC. Multiple myeloma: the (r)evolution of current therapy and a glance into future. Haematologica. 2020;105(10):2358-2367.
doi pubmed - Kumar S, Fonseca R, Ketterling RP, Dispenzieri A, Lacy MQ, Gertz MA, Hayman SR, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012;119(9):2100-2105.
doi pubmed - Neben K, Jauch A, Hielscher T, Hillengass J, Lehners N, Seckinger A, Granzow M, et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J Clin Oncol. 2013;31(34):4325-4332.
doi pubmed - Rajkumar SV, Gupta V, Fonseca R, Dispenzieri A, Gonsalves WI, Larson D, Ketterling RP, et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia. 2013;27(8):1738-1744.
doi pubmed - Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95(5):548-567.
doi pubmed - Kwon WK, Lee JY, Mun YC, Seong CM, Chung WS, Huh J. Clinical utility of FISH analysis in addition to G-banded karyotype in hematologic malignancies and proposal of a practical approach. Korean J Hematol. 2010;45(3):171-176.
doi pubmed - Shin SY, Eom HS, Sohn JY, Lee H, Park B, Joo J, Jang JH, et al. Prognostic implications of monosomies in patients with multiple myeloma. Clin Lymphoma Myeloma Leuk. 2017;17(3):159-164.e152.
doi pubmed - Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, Attal M, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27(27):4585-4590.
doi pubmed - Cardona-Benavides IJ, de Ramon C, Gutierrez NC. Genetic abnormalities in multiple myeloma: prognostic and therapeutic implications. Cells. 2021;10(2):336.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


