| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website https://www.thejh.org |
Short Communication
Volume 13, Number 3, June 2024, pages 86-93
Frameshift Mutations in Leukemia-Associated Genes Correlate With Superior Outcomes in Patients Undergoing Allogeneic Stem Cell Transplant for De Novo Acute Myeloid Leukemia
Emma Cammanna, Sindha Madhavb, Lloyd Hutchinsonb, Jan Cernyc, Muthalagu Ramanathanc, Jacob R. Bledsoed, Vladislav Makarenkoe, Shyam A. Patelc, Xiuling Mengb, Keith Tomaszewiczb, Rajneesh Nathf, Benjamin Cheng, Bruce Wodab, William Selovea, h, i
aUMass Medical School, Worcester, MA, USA
bDepartment of Pathology, UMass Memorial Medical Center, University of Massachusetts, Worcester, MA, USA
cDepartment of Hematology-Oncology, UMass Memorial Medical Center, University of Massachusetts, Worcester, MA, USA
dDepartment of Pathology, Boston Children’s Hospital, Boston, MA, USA
eCommonwealth Pathology Partners, P.C., Salem, MA, USA
fDepartment of Hematology, Medical Oncology, Banner MD Anderson Cancer Center Clinic, Gilbert, AZ, USA
gBristol Myers Squibb Company, Cambridge, MA, USA
hDepartment of Pathology, Baystate Medical Center, Springfield, MA, USA
iCorresponding Author: William Selove, Department of Pathology, Baystate Medical Center, Springfield, MA, USA
Manuscript submitted April 13, 2024, accepted May 28, 2024, published online June 28, 2024
Short title: Frameshift Mutations and AML Survival Posttransplant
doi: https://doi.org/10.14740/jh1276
| Abstract | ▴Top |
Background: Allogeneic stem cell transplant (allo-SCT) is a mainstay of treatment for acute myeloid leukemia (AML). Its success depends largely on response of donor T lymphocytes against leukemia cells, known as graft-vs-leukemia (GvL) effect. A key potential driver of GvL is immune response to mutation-derived neoantigens. Previous studies in solid tumors have demonstrated enhanced immunogenicity of frameshift (FS)-derived peptides vs. those from non-synonymous single nucleotide variants (SNVs). We therefore hypothesized that AML cases bearing FS mutations in leukemia-associated genes would be more immunogenic than those with only other types of mutations (non-FS), and thus benefit more from allo-SCT via more robust GvL.
Methods: We identified AML patients who had undergone allo-SCT between 2010 and 2022 and had next-generation sequencing data available on diagnostic specimens using a 42-gene hot spot panel. We compared the impact of tumor mutations present at diagnosis on overall survival and relapse-free survival based on FS versus non-FS status.
Results: Ninety-five AML allo-SCT patients were identified. We observed superior relapse-free survival (P = 0.038, hazard ratio (HR): 0.24) and borderline superior overall survival (P = 0.058, HR: 0.55) post-transplant in de novo AML patients, who had at least one FS mutation (other than NPM1) in one of the 42 assessed genes versus those with only non-FS mutations.
Conclusions: Our findings suggest that FS-mutated AML cases may benefit more from allo-SCT than those with only non-FS mutations, possibly due to increased generation of immunogenic neoepitopes. If validated in an expanded study, incorporation of somatic FS mutation status in AML could improve patient selection algorithms for bone marrow transplant and thereby lead to superior outcomes.
Keywords: AML; Transplant; Frameshift; NGS; Survival; Allogeneic; TMB
| Introduction | ▴Top |
Acute myeloid leukemia (AML) can be a rapidly fatal disease for which allogeneic stem cell transplant (allo-SCT) offers the only chance of cure. The success of allo-SCT, in which a patient’s hematopoietic stem cells are replaced with those of an allogeneic donor, hinges in great part on the immune response of donor T lymphocytes against leukemia cells (graft-vs-leukemia (GvL) effect). Unfortunately, donor T cells can also damage non-leukemic tissue in the host (graft-vs-host disease (GvHD)) [1]. This raises the question: Which specific leukemia cases have characteristics that make them maximally vulnerable to GvL effect while minimizing GvHD?
The answer to this question may be informed by the three types of antigens against which donor T lymphocytes act. These are: major histocompatibility antigens, minor histocompatibility antigens, and cancer-specific neoepitopes [2]. Major histocompatibility antigens and minor histocompatibility antigens are present on both host cells and neoplastic cells and therefore serve as targets for both GvHD and GvL effects. Cancer-specific neoepitopes, on the other hand, are present only on the neoplastic cells, so immune response against these antigens would almost exclusively target tumor cells while producing relatively little damage to non-neoplastic host tissue (thereby avoiding on-target, off-tumor toxicity). It would seem then that neoepitope-rich leukemias should benefit from relatively greater GvL-specific effect than cancers with relatively fewer neoepitopes.
There are several mutational mechanisms that may generate neoepitopes. These include copy number variations, translocations, nonsense mutations giving rise to stop codons, missense mutations, and frameshift (FS) mutations. Any of these mutations can incite immune attack through altered protein expression, but they are not all equivalent in their ability to do so. Consider a missense mutation for instance. A missense causes a substitution in just a single amino acid and would therefore have a relatively low probability of causing the display of an immunogenic peptide. FS mutations by contrast can generate long novel peptide strings distal to the site of mutation. The amino acid sequence in these peptides would of course be “foreign” and would contain many potentially immunogenic targets. Hence, one would expect FS mutations to induce a more robust immune response than other types of mutations. In fact, previous studies in solid tumors have demonstrated the enhanced immunogenicity of FS-derived proteins vs. others in various cancer types [3-5]. Sena et al for example established much stronger correlation of FS proportion than overall tumor mutation burden (TMB) for response of prostate cancer to programmed death ligand-1 (PD-L1) inhibitors [6].
We might therefore predict that AML cases bearing relatively higher numbers of FS mutations would be more immunogenic than those with fewer and thus be more responsive to treatment with allo-SCT through more vigorous GvL effect. We sought to test this hypothesis by exploring the impact of FS mutations (≥ 1) as detected by a hot spot next-generation sequencing (NGS) panel (42 total genes in the currently used test) on post-transplant outcomes. As at other institutions, NGS to assess for mutations in commonly altered oncogenes and tumor suppressors is routinely performed on diagnostic and follow-up specimens of patients with AML at UMass. We retrospectively reviewed our electronic medical record (EMR) for patients who had undergone allo-SCT for AML and assessed the impact of tumor mutations present at diagnosis on overall survival (OS) and relapse-free survival (RFS), based on FS versus no FS status to determine whether the former would preferentially benefit more from allo-SCT.
| Materials and Methods | ▴Top |
This study protocol was reviewed and approved by the UMass Medical School Institutional Review Board (IRB) (approval number 0014). This study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
With UMass IRB approval, we searched our institutional EMR for patients with AML, who had undergone allo-SCT between 2009 and 2022 and had NGS data available on diagnostic specimens using a 42-gene hot spot panel. Diagnostic bone marrow or peripheral blood specimens were sequenced via the CTMP version 3, 42 gene myeloid panel on an Ion Torrent PGM next-generation sequencer. Somatic mutations in diagnostic specimens were called based on tier 1 (pathogenic) or tier 2 (likely pathogenic) mutations at variant allele frequencies (VAFs) ≥ 2%. NPM1 and FLT3 mutation status was tested in the diagnostic specimens of all patients using polymerase chain reaction (PCR) fragment analysis. Via the Kaplan-Meier method, we analyzed and compared the impact of tumor mutations present at diagnosis on OS and RFS based on FS versus no FS status. The FS group was defined as having at least one frameshifted gene other than NPM1. The non-FS group had no non-NPM1 FSs in any of the 42 assessed genes. Relapse was defined by more than 5% bone marrow blasts or reappearance of circulating blasts after a documented complete remission. OS was defined as the interval between day of allo-SCT and day of death or last follow-up. α was set at 0.05 to assess for statistical significance.
| Results | ▴Top |
We identified 95 patients with AML (patient and transplant characteristics were shown in Table 1) who had at least one mutation detected by NGS or PCR fragment analysis (for FLT3-ITD and NPM1). Frameshifted genes identified included RUNX1, WT1, TP53, BCOR, CEBPA, ETV6, STAG2, PHF6, ZRSR2, TET2, EZH2, ASXL1 and NPM1 (Table 2). Non-FS mutations included RUNX1, TET2, BCOR, ASXL1, WT1, STAG2, ETV6, ZRSR2, CEBPA, TP53, PHF6, EZH2, FLT3, NRAS, DNMT3A, IDH2, IDH1, PTPN11, SRSF2, CBL, U2AF1, SETBP1, JAK2, SF3B1, JAK3, KIT. Patients included 78 with de novo leukemia and 17 with secondary AML (nine in the FS group and eight in the non-FS group (P = 0.25, not significant (NS)). FS patients showed a significantly higher proportion of National Comprehensive Cancer Network (NCCN)-defined adverse risk disease than non-FS patients (P = 0.011). TP53 mutations specifically included two FSs and nine non-FSs (P = 0.20, NS). Thirty-four patients had at least one gene bearing exclusively FS mutations. Fifty-seven patients had no FS other than NPM1. Four patients with FSs had overlapping non-synonymous single nucleotide variants (SNVs) in all identified frameshifted genes. This last group was excluded from analysis. The median age was 65 (interquartile range (IQR): 55 - 72) years in the FS group and 62 (IQR: 49 - 69) in the non-FS group. Median follow-up time for the FS group was 79.4 months, and 74.9 months for the non-FS group. Among patients for whom GvHD data were available (n = 66), acute or chronic GvHD occurred at similar frequencies in the non-FS (n = 18/38, 47%) and FS (n = 15/28, 54%) groups (P = 0.62).
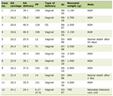 Click to view | Table 1. Demographics and Disease Characteristics |
 Click to view | Table 2. Mutated Genes at Diagnosis With Respective Numbers of Frameshift and Non-Frameshift Variants |
Survival of all patients (including both primary and secondary AML)
Median OS was 72.1 months for patients who had at least one exclusively frameshifted gene (n = 34) other than NPM1 vs. 12.9 months for other patients (n = 57). On univariate analysis, the hazard ratio (HR) for OS was 0.59 (confidence interval (CI): 0.35 - 0.97, P = 0.042) in favor of the frameshifted group (Table 3, Fig. 1). The HR for RFS was 0.55 (CI: 0.23 - 1.30) which was not significant (P = 0.20) (Fig. 2). Multivariate analysis (Table 4) extends the significance of the OS advantage in the FS group (HR: 0.49 (0.26 - 0.87), P = 0.018). RFS in this all-inclusive group (primary and secondary) after adjusting for relevant factors (including age, NCCN biological risk category, de novo vs. secondary disease, presence/absence of GvHD) was still not significant but approached more closely (HR: 0.42 (0.16 - 1.09), P = 0.08). Additionally of note, secondary AML showed significantly higher risk for mortality (2.3 (1.18 - 4.46), P = 0.014) and relapse (4.12 (0.51 - 12.3), P = 0.006) than de novo AML in our cohort.
 Click to view | Table 3. Outcome Summary: FS vs. No-FS (Univariate Analysis) |
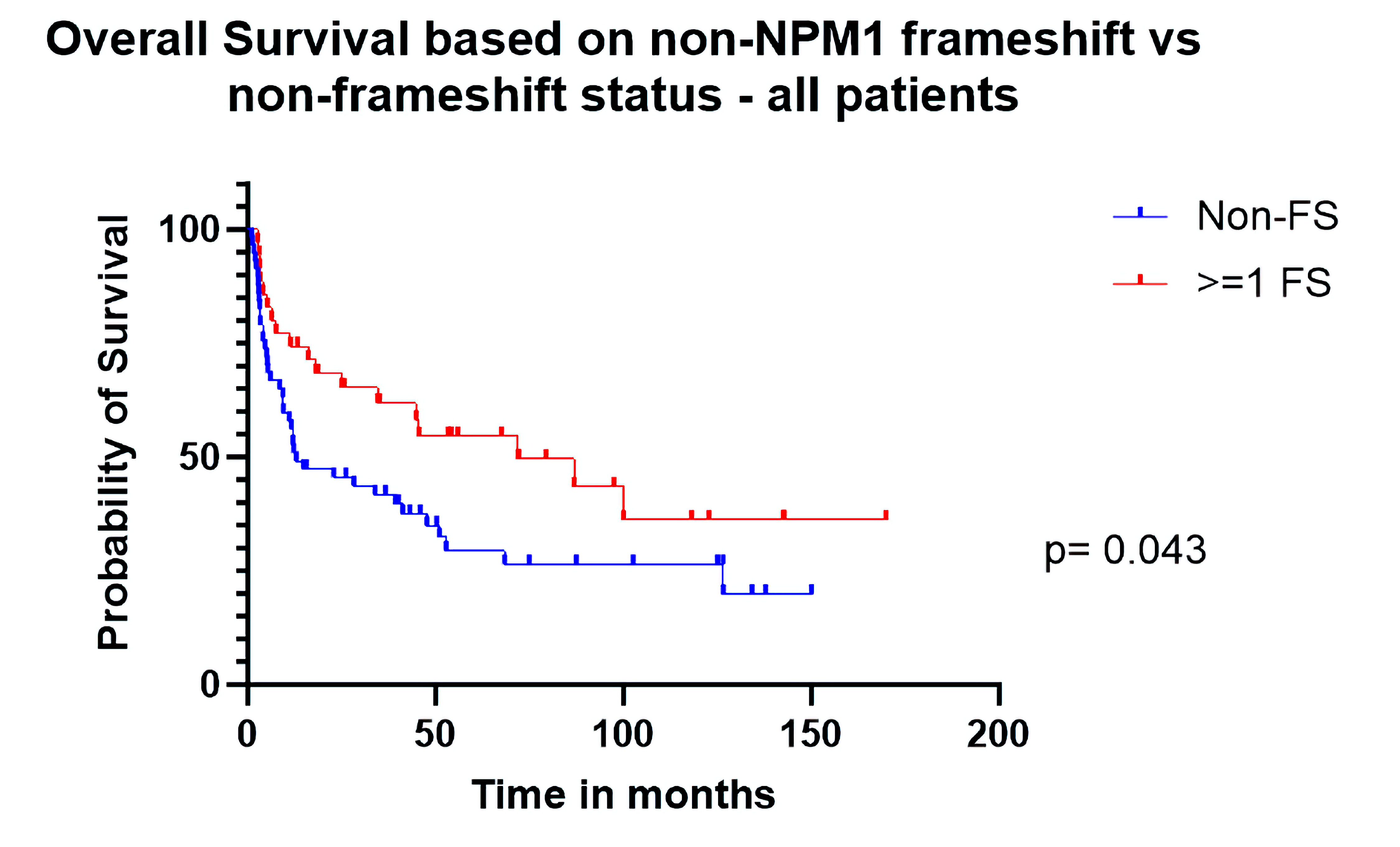 Click for large image | Figure 1. Overall survival based on non-NPM1 frameshift (FS) vs. non-FS status (all patients). |
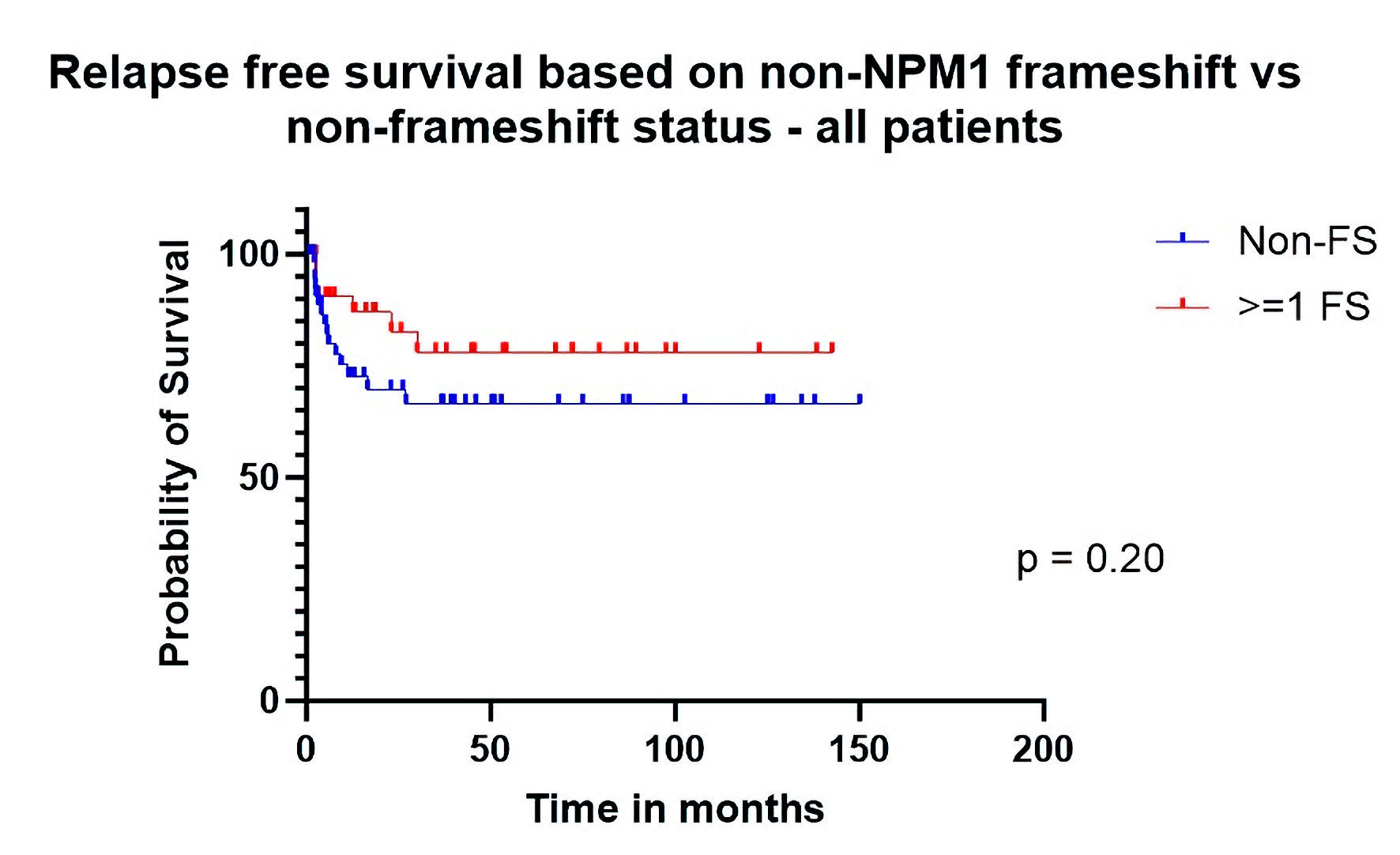 Click for large image | Figure 2. Relapse-free survival based on non-NPM1 frameshift (FS) vs. non-FS status (all patients). |
 Click to view | Table 4. Outcome and Multivariate Analysis of Relevant Factors |
Survival among patients with de novo AML
When analysis was restricted to patients with de novo disease, post-transplant RFS was significantly better (HR: 0.24 (CI: 0.08 - 0.68), P = 0.039) in the FS group (Fig. 3). Median OS was 100 months for patients who had at least one gene with FS mutations (n = 26) other than NPM1 vs. 33 months for other patients (n = 49). The HR for OS was 0.55 (CI: 0.30 - 1.02) with borderline significance (P = 0.058) (Fig. 4) in univariate analysis. When adjusting for relevant factors with multivariate analysis, both RFS (HR: 0.16 (0.03 - 0.79), P = 0.024) and OS (HR: 0.45 (0.22 - 0.92), P = 0.028) were significantly better in the FS group.
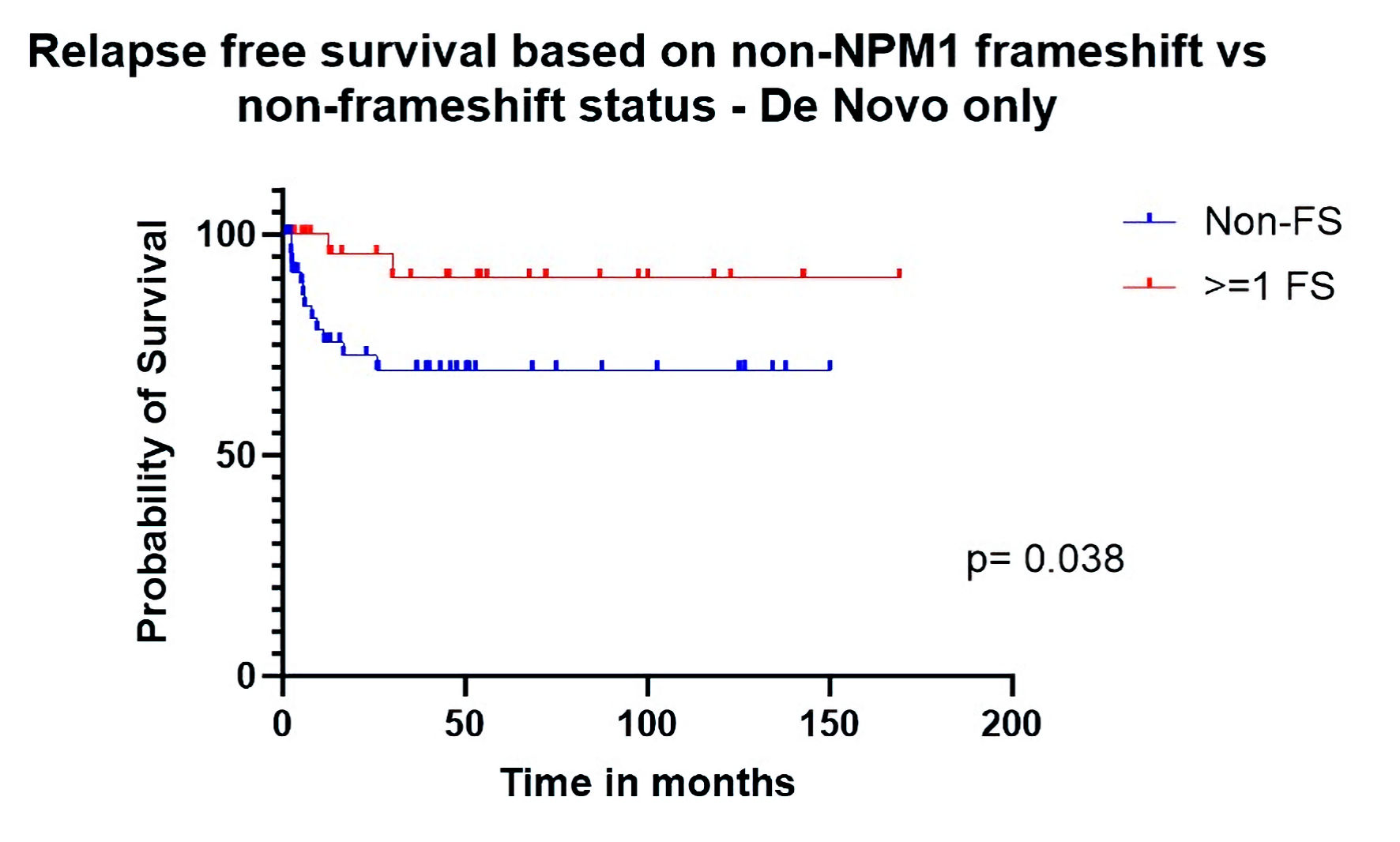 Click for large image | Figure 3. Relapse-free survival based on non-NPM1 frameshift (FS) vs. non-FS status (de novo only). |
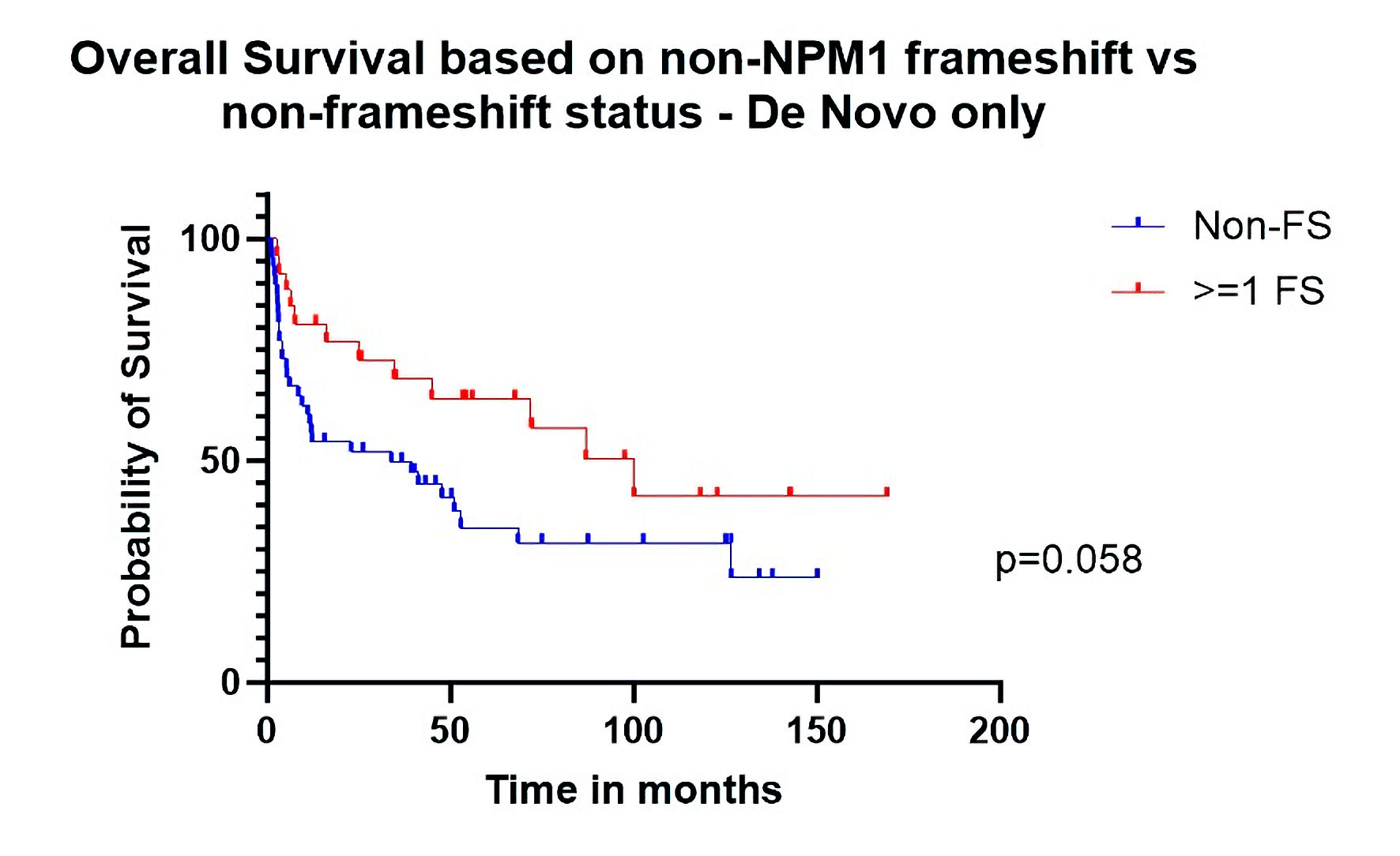 Click for large image | Figure 4. Overall survival based on non-NPM1 frameshift (FS) vs. non-FS status (de novo only). |
Survival of RUNX1 FS vs. non-FS
Additionally, among de novo AML patients with RUNX1 mutations (n = 16) specifically, those with frameshifted RUNX1 (n = 11) demonstrated superior overall post-transplant survival (HR: 0.21, P = 0.042) to those who harbored non-frameshifted RUNX1 (n = 5) (Fig. 5). Other mutations occurred with insufficient frequency to individually compare FS vs. non-FS survival effects at the level of single genes.
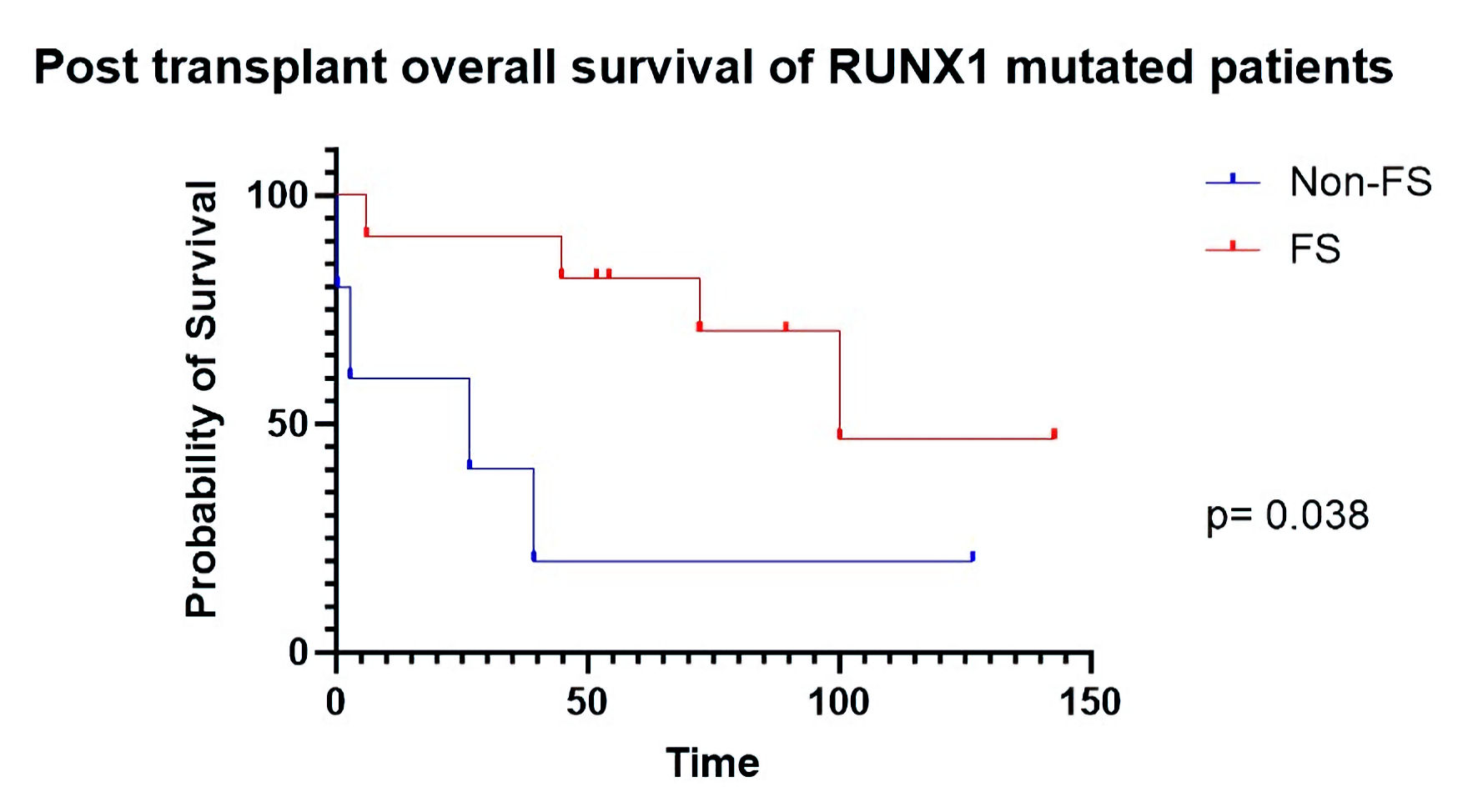 Click for large image | Figure 5. Post-transplant overall survival of RUNX1 mutated patients. FS: frameshift. |
| Discussion | ▴Top |
In summary, in our cohort, among all AML transplant patients, those with at least one FS mutation other than NPM1 showed superior post-transplant OS and borderline superior RFS to those with other types of mutations. Further, among patients with de novo AML, in addition to superior OS, frameshifted cases demonstrated significantly longer RFS. Moreover, these survival advantages come in spite of the fact that the FS group in our cohort includes a significantly higher proportion of NCCN-defined biologically adverse risk cases.
The evolutionary fitness of cancer can be crudely viewed as directly proportional to its ability to generate diversity through mutations and inversely proportional to its propensity to attract the attention of T cell “predators” of the immune system. For malignant cells, FS mutations are an effective means of knocking out tumor suppressors but could increase vulnerability to immune therapy. Turajlic et al have demonstrated that FS indel mutations have on average nine times the immunogenicity of SNV mutations [4, 7]. Hence higher proportions of FS mutations, bearing more mean immunogenic potential than SNV, might tip the scales in favor of susceptibility to immune therapy. In fact, increased OS and PFS have been previously correlated with higher FS proportions in prostate carcinoma and clear cell renal cell carcinoma patients on PD-L1 therapy [4, 6]. Allo-SCT is of course in great part an immune therapy. It exploits the fact that donor T cells, unlike recipient T cells, are not inured to the presence of leukemia and will aggressively seek out neoplastic cells. FSs generally should make AML more easily recognizable than SNV, facilitating leukemia eradication. Our findings appear to support this theory, demonstrating a novel correlation between somatic FS mutations and post-transplant survival for patients with AML.
Additionally of interest, FS cases in our cohort demonstrated similar frequency of GvHD to non-FS cases (P = 0.62). This is as would be expected since according to our theory, augmented immunity in the FS cases would be neo-antigen specific and would have no impact on immune response against host tissue.
TMB is of course well known to correlate with response to immune therapy in patients with solid tumors and is classically calculated using whole exome sequencing (WES). WES is not practical outside of the research setting, but targeted gene panels have been used as an effective surrogate for establishing TMB in solid tumors [1, 8]. Similar targeted NGS panels are already commonly used clinically in the diagnosis and characterization of AML [9]. These tests help define specific World Health Organization-defined or International Consensus Classification (ICC)-defined entities (e.g., AML with mutated NPM1), identify therapeutic targets (e.g., FLT3 and IDH1/2) or help establish the presence of residual disease post-treatment. In current practice, with respect to AML, however, results of NGS are interpreted solely with an eye to determining whether a mutation is likely to be pathogenic rather than to establish whether or not a mutation may be immunogenic. Hence, in the context of AML, FS mutations, substitutions, stop codons, etc., are all viewed as more or less equivalent as long as they have a meaningful impact on oncogenesis. Vulnerabilities that such mutations may cause through enhanced immune recognition are largely ignored, and there is relatively little literature on the impact of TMB, much less FS burden, on allo-SCT outcomes in AML. Foran et al did not note a correlation of TMB in allo-SCT with post-transplant outcomes. However, their study would have benefitted from a higher sample size and evaluation of FS burden [7]. Results of routine clinical NGS testing represent an untapped trove of data with respect to potential immunogenic targets in AML.
FS mutations are clearly a heterogeneous group, and questions remain with respect to peptide length prior to a stop codon, role of nonsense-mediated decay and likelihood of peptide display by donor major histocompatibility complex (MHC) molecules in individual cases. One major question our study raises is why patients with FS mutations other than NPM1 generally fared better than those with only NPM1 FS (HR: 0.57) though not significant (P = 0.12) in our cohort (Supplementary Material 1, www.thejh.org). NPM1 aberration in the vast majority of cases shows the same 4-bp insertion, thought to be mediated by mistargeted base pair insertion by terminal deoxynucleotidyl transferase (TdT) [10, 11]. Prior studies have shown IFN-γ-producing NPM1-mutated-specific T cells in PB samples obtained in the allo-SCT setting. Importantly, however, human leukocyte antigen (HLA) class I alleles, such as B*07, B*18, and B*40, predicted to bind potent NPM1-derived immunopeptides, show a reduced incidence in the NPM1-mutated AML cases compared to healthy subjects and NPM1-wild-type AML patients [12-14]. Taken together, these findings suggest that despite potential NPM1 immunogenicity, NPM1-positive allo-SCT transplant cases may be enriched for patients who lack the HLA molecules necessary to present them. Further, it may be that the product of the NPM1 FS is not on average as immunogenic as other FS mutations are.
Further investigation is certainly required. Mechanistic investigations are needed to establish whether the superior outcomes we observed in patients with FSs are indeed due to immunogenic response to neoepitopes as hypothesized. Immunopeptidome studies to evaluate the capability of host antigen presenting cells to present the leukemia-derived neoantigens and donor T cells to recognize them would be of great interest. Our study is also limited by relatively small sample size and incomplete data regarding disease status and patient performance status at the time of transplant. Still, our data suggest that patients with AML that harbors FS mutations may benefit more from allo-SCT than those with only non-FS mutations, possibly due to increased generation of immunogenic neoepitopes. If validated in larger studies, incorporation of somatic FS mutation status in AML risk stratification models could help identify patients in whom allo-SCT would be maximally beneficial.
| Supplementary Material | ▴Top |
Suppl 1. Overall survival based on frameshift status (all patients).
Acknowledgments
None to declare.
Conflict of Interest
M. Ramanathan: Spouse owns stock options in Smith & Nephew: Current equity holder in publicly-traded company; spouse owns stocks in Alcon, Biogen, Boston Scientific, Dexcom, Gilead, Intuitive Surgical, Moderna, Medtronic, Novartis, Tandem Diabetes, Ocular Therapeutics, Pfizer, Sanofi, Vertex, and Viatris: Current equity holder in publicly-traded company; spouse has divested stocks in Abbvie, Jansenn and BMS: Divested equity in a private or publicly-traded company in the past 24 months. J. Cerny: MERIT CRO: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Actinium Pharmaceuticals: Current holder of stock options in a privately-held company; BlueBird Bio, Inc.: Current holder of stock options in a privately-held company; Cellectar Sciences: Current holder of stock options in a privately-held company; Dynavax Technologies: Current holder of stock options in a privately-held company; Gamida Cell: Current holder of stock options in a privately-held company; Atyr Pharma: Current holder of stock options in a privately-held company; Novavax Inc.: Current holder of stock options in a privately-held company; Ovid Therapeutics Inc.: Current holder of stock options in a privately-held company; Sorreto Therapeutics: Current holder of stock options in a privately-held company; Veru Inc.: Current holder of stock options in a privately-held company; Viridian Therapeutics: Current holder of stock options in a privately-held company; Vaxart Inc.: Current holder of stock options in a privately-held company; 2Seventy Bio Reg SHS: Current holder of stock options in a privately-held company; Bristol Myers Squibb: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; ICON-Prolacta: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; ICON-Allovir: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees. S.A. Patel: UMass Center for Clinical and Translational Science (CCTS) Pilot Project Program grant (NIH/NCATS Grant UL1TR001453: Research Funding; Bristol Myers Squibb: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Pfizer: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees.
Informed Consent
Written informed consent was not required, per exemption from the IRB above.
Author Contributions
William Selove conceived and designed the study. Rajneesh Nath, Muthalagu Ramanathan, Jan Cerny and Shyam A. Patel contributed to clinical data. Emma Cammann, William Selove, Sindha Madhav, Vladislav Makarenko populated the data base with NGS and clinical findings. Emma Cammann, William Selove, Lloyd Hutchinson, Xiuling Meng, Keith Tomaszewicz, Vladislav Makarenko, and Jacob R. Bledsoe analyzed the data. William Selove and Emma Cammann wrote the paper. Bruce Woda, Lloyd Hutchinson, Muthalagu Ramanathan, Benjamin Chen, Jan Cerny and Shyam A. Patel reviewed and revised the paper.
Data Availability
The data that support the findings of this study are available from the corresponding author, WJS, upon request.
| References | ▴Top |
- Tabbara IA, Zimmerman K, Morgan C, Nahleh Z. Allogeneic hematopoietic stem cell transplantation: complications and results. Arch Intern Med. 2002;162(14):1558-1566.
doi pubmed - Sweeney C, Vyas P. The Graft-Versus-Leukemia Effect in AML. Front Oncol. 2019;9:1217.
doi pubmed pmc - Schwitalle Y, Linnebacher M, Ripberger E, Gebert J, von Knebel Doeberitz M. Immunogenic peptides generated by frameshift mutations in DNA mismatch repair-deficient cancer cells. Cancer Immun. 2004;4:14.
pubmed - Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong YNS, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18(8):1009-1021.
doi pubmed - Litchfield K, Reading JL, Lim EL, Xu H, Liu P, Al-Bakir M, Wong YNS, et al. Escape from nonsense-mediated decay associates with anti-tumor immunogenicity. Nat Commun. 2020;11(1):3800.
doi pubmed pmc - Sena LA, Fountain J, Isaacsson Velho P, Lim SJ, Wang H, Nizialek E, Rathi N, et al. Tumor frameshift mutation proportion predicts response to immunotherapy in mismatch repair-deficient prostate cancer. Oncologist. 2021;26(2):e270-e278.
doi pubmed pmc - Foran JM, Heckman MG, Cherukuri Y, Wieczorek M, Abdel Rahman Z, Jiang L, Murthy HS, et al. Epidemiologic and clinical analysis of Tumor Mutational Burden (TMB) in Acute Myeloid Leukemia (AML): exome sequencing study of the Mayo Clinic AML Epidemiology Cohort (MCAEC). Blood. 2021;138:3437.
- Fancello L, Gandini S, Pelicci PG, Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer. 2019;7(1):183.
doi pubmed pmc - Leisch M, Jansko B, Zaborsky N, Greil R, Pleyer L. Next generation sequencing in AML-on the way to becoming a new standard for treatment initiation and/or modulation? Cancers (Basel). 2019;11(2):5020.
doi pubmed pmc - Borrow J, Dyer SA, Akiki S, Griffiths MJ. Molecular roulette: nucleophosmin mutations in AML are orchestrated through N-nucleotide addition by TdT. Blood. 2019;134(25):2291-2303.
doi pubmed - Zarka J, Short NJ, Kanagal-Shamanna R, Issa GC. Nucleophosmin 1 mutations in acute myeloid leukemia. Genes (Basel). 2020;11(6):649.
doi pubmed pmc - Greiner J, Schneider V, Schmitt M, Gotz M, Dohner K, Wiesneth M, Dohner H, et al. Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut). Blood. 2013;122(6):1087-1088.
doi pubmed - Kuzelova K, Brodska B, Fuchs O, Dobrovolna M, Soukup P, Cetkovsky P. Altered HLA Class I Profile Associated with Type A/D Nucleophosmin Mutation Points to Possible Anti-Nucleophosmin Immune Response in Acute Myeloid Leukemia. PLoS One. 2015;10(5):e0127637.
doi pubmed pmc - Forghieri F, Riva G, Lagreca I, Barozzi P, Bettelli F, Paolini A, Nasillo V, et al. Neoantigen-specific T-cell immune responses: the paradigm of NPM1-mutated acute myeloid leukemia. Int J Mol Sci. 2021;22(17):9159.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


