| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website http://www.thejh.org |
Original Article
Volume 7, Number 1, January 2018, pages 7-13
Role of Nucleophosmin Gene Mutation in Leukemogenesis of Acute Myeloid Leukemia
Safaa A.A. Khaleda, e, John Burthemb, El-Badry E. Abo Elnoorc, Lobna F. ElTonic, Hanan M. Ahmedc, Sohier M. Ahmedd
aDepartment of Internal Medicine, Hematology and BMT Unit, Assiut University Hospital, Assiut 71111, Egypt
bDepartment of Hematology, Manchester Royal Infirmary, Manchester School of Medicine, Manchester University, UK
cDepartment of Internal Medicine, Assiut University Hospital, Faculty of Medicine, Assiut University, Egypt
dDepartment of Clinical Pathology, Assiut University Hospital, Faculty of Medicine, Assiut University, Egypt
eCorresponding Author: Safaa A. A. Khaled, Department of Internal Medicine, Hematology and BMT Unit, Assiut University Hospital, Assiut 71111, Egypt
Manuscript submitted December 4, 2017, accepted December 15, 2017
Short title: Prolliferative Role of Mutated NPM in AML
doi: https://doi.org/10.14740/jh365w
| Abstract | ▴Top |
Background: Acute myeloid leukemia (AML) is a hematopoietic stem cell disorder that carries very poor prognosis. Understanding molecular basis of AML leukemogenesis could lead to the emergence of effective targeted therapies for AML. AML bearing nucleophosmin (NPM) gene mutation has distinct features. This study was conducted to investigate the role of mutated (m) NPM in pathogenesis of de novo AML through studying its contribution in proliferation of AML cell line cells.
Methods: Two types of human leukemia cell lines were used. One of them was a model for AMLs with mNPM and the other for AMLs with wild type (wt) NPM. Assessment of the proliferative role of mNPM in AML was carried out using cell culture and viability studies. The obtained results were reaffirmed by immunocytochemical and immunoblotting techniques.
Results: Analysis of results was done with the appropriate computer software. It showed higher proliferative potential of cells with mNPM compared to those bearing wtNPM only. Furthermore, the immunocytochemical studies demonstrated subcellular localization of NPM isoforms during various phases of mitosis. Mitosis was associated with cytoplasmic translocation of wtNPM in certain phases, while localization of mNPM remained unchanged throughout the cell cycle. Results of immunoblotting showed little or no change in protein expression of either NPM moieties during mitosis.
Conclusions: The current study demonstrated important contribution of NPM gene mutation in enhancing proliferation of AML cell lines. These results confirmed the role of mNPM in AML leukemogenesis, and highlighted the importance of targeting mNPM in new evolving AML therapies.
Keywords: AML; Nucleophosmin; Leukemogenesis; Proliferation
| Introduction | ▴Top |
Cancer usually results from the disruption of the proliferation/apoptosis cellular balance. A recent approach in cancer management is to create anticancer drugs that could change the molecular and biological properties of cancer cells and return the balance to normal [1]. Acute myeloid leukemia (AML) is a cytogenetically and clinically heterogeneous disease [2], recent studies revealed the presence of nucleophosmin (NPM) gene mutation in 65% of de novo AML with normal karyotype [3]. In respect to this high incidence of NPM mutation in AML it is conceivable that mutated (m) NPM plays an important role in AML leukemogenesis and development. Also AML bearing mNPM had been included as provisional entity in the World Health Organization classification (WHO) in 2008. According to the WHO AML with mNPM is termed NPMc + AML, referring to the aberrant cytoplasmic localization of mNPM [4].
A variety of laboratory techniques could be used to study mNPM in AML cells. Immunocytochemistry (ICC) is a laboratory technique based on the antigen-antibody interaction. ICC could be used to study mNPM on AML; also it allows analysis of NPM behavior throughout the cell cycle. This is conducted by direct microscopic examination of AML cytospins immunostained with NPM. Under microscopy, the morphological appearance of cells will help track NPM during mitosis [5]. Immunoblotting is another laboratory technique that allows studying NPM [6], it is a procedure that helps not only to identify an antigen but also to determine its MW, quantity and possible protein interactions. This is achieved by binding the protein obtained by gel electrophoresis with the antigen of interest with specific antibodies to the antigen [7].
In this study mNPM was meticulously investigated during proliferation of AML cells, aiming to study its pathologic role in primary AML. The study was conducted on both NPM moieties (wild type (wt) NPM and mNPM) using type A mNPM as an example for mNPM.
| Materials and Methods | ▴Top |
Some technical and ethical reasons limited this study to be in vitro study only. Accordingly, human leukemia cell lines were used as a model for AML cells. Accordingly two types of human leukemia cell lines were used in this work, one to act as a model for NPMc + AML (AML bearing mNPM) and the other as a model for NPMc-AML (AML with mild type AML). In order to investigate the role of mNPM in proliferation of AML cells the behavior of both NPM moieties was studied throughout mitosis of colchicine treated cells of both cell types.
Biological materials
Human leukemia cell lines
Two human leukemia cell lines were used, OCI-AML3 Cells were purchased from DSMZ (German collection of micro-organisms and culture) (ACC, 582). Mutational analysis of NPM in these cells revealed TCTG duplication at 956-959 of exon 12- of NPM gene (GenBank accession number NPM-002520). This corresponds to type A NPM mutation that was found in NPMc + AML (NPM-cytoplasmic positive) [8]. Accordingly, OCI-AML3 was used as a cell line model for NPMc+AML.
HL60 cells were purchased from DSMZ (ACC, 3); we used HL60 as a cell line model for NPMc- AML (NPM-cytoplasmic negative).
Antibodies
Two types of antibodies were used, primary and secondary antibodies. Primary antibodies include NPM FC-61991, from Zymed which is specific to NPM C-terminus region, NPM Ab 15440 from Abcam that recognizes NPM a.a.23-38 and 226-240, and NPM NA24 from Thermo that specifically reacting with NPM N-terminal region and 14a.a ALK.
A panel of species specific fluorophore conjugated secondary antibodies was used. These antibodies are Texas Red (TR) conjugated (Abcam 6003), and fluorescence isothiocyanate (FITC) conjugated (Abcam 6717). An enzyme linked secondary antibody was used in proteomic analysis that was obtained from GE-Healthcare.
Other materials
Other materials like chemicals, equipments, instruments and computer software will be mentioned together with the methods.
Methods
Cell culture and viability studies
Cells were grown in RPMI media 1640, with GlutaMAX™ supplemented with 10% FCS and 1%Pen/strep (GIBCO, 61870-044, 10108-165, and15070-063). Cells were freshly fed, then each culture was divided into two flasks, one flask was left to grow normally (control), 1µM colchicine was added to the other. Colchicine was used at a dilution 1:100 and incubated for 24 h. After 4 and 24 h cells were extracted from the media, of the control and treated cultures, cellular cytospins and lysates were prepared. Cytospins were used to assess the proliferative potential of each culture, through determination of the percent of blocked metaphases, also to investigate the kinetic changes in NPM during various stages of cell cycle. On the other hand the whole cellular lysates (WHL) were used to assess changes in NPM protein expression during cellular proliferation.
Immunoblot analysis
Cells were pelleted, washed, and lysed by incubation with RIPA buffer. Lysates were centrifuged and the supernatant was collected, diluted then loaded into 12% SDS-PAGE gels. Next it was electroblotted with the same protocol described by others in 1979 [9], with some modifications, such as the use of polyvinylidene fluoride (PVDF) membrane instead of the nitrocellulose membrane. The obtained protein bands were blotted with a panel of anti-human NPM antibodies. Lastly, images of the obtained blot were captured using a Kodak X-Ray film and cassette (Sigma Aldrich, Kodak Biomax XAR Film size 18/24 cm). Films were developed and the obtained protein bands were then marked compared to the Bio-Rad Precision Protein Standards.
Immunocytochemistry (Immunostaining)
For preparation of cytospins from both cultures, a volume of 100 - 200 µL containing a number of 5 × 104 to 2 × 105 viable cells was added in a pre-assembled cytospin buckets including frosted slides (SLS, MIC3806).Then the buckets were spun at 60 × 10 for 5 min with the acceleration off in a specific cytospin machine (Shandon). Buckets were dismantled, and the slides left to dry at RT overnight. Lastly, the slides were wrapped in a foil and freezed at -8 °C. The cytospin quality was assessed by mounting the cytospin in a drop of reagent with DAPI (Invitrogen, P36935), then viewed under the immunofluorescence microscope.
We followed the manufacturer recommended protocols for immunostaining of cytospins for NPM. After which, the slides were viewed, with the Nikon Eclipse 80I fluorescence microscope (ICCT technologies, Canada), equipped with three fluorescence filter cubes. The obtained images were grabbed with the Hamamatsu ORCA HR (C4742-95-12HR) high resolution digital camera with full remote control from PC. Fast high sensitivity black and white camera was used for low intensity fluorescence slides. Analysis of the obtained images was conducted with the NIS-elements software.
| Results | ▴Top |
Colchicine effect on AML derived cells
Examination of the 4 h colchicine treated cytospins after mounting in DAPI to stain the nuclei, showed a good demonstration of most of the cell cycle stages, as depicted in Figure 1, besides the blocked metaphases. This indicated that a fraction of the cells entered metaphase and completed mitosis but this fraction is expected to decrease with time until a complete block achieved.
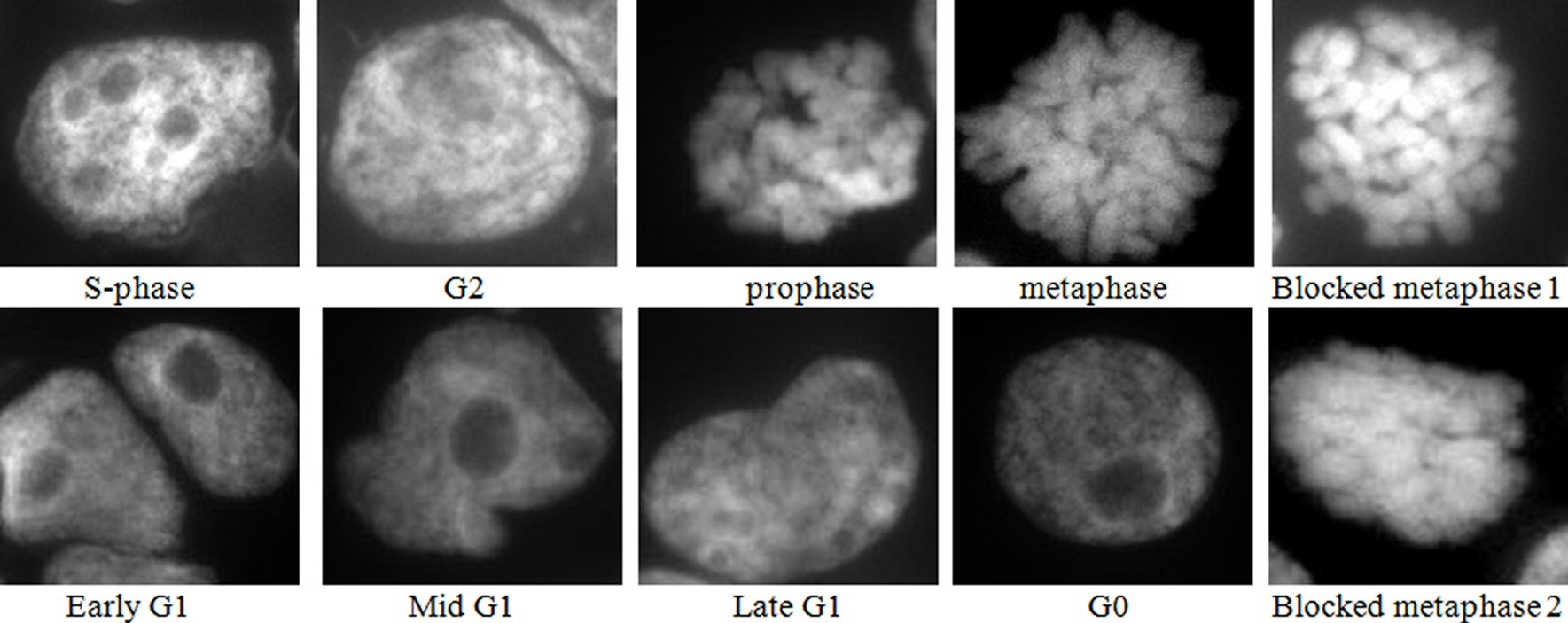 Click for large image | Figure 1. Images of DAPI mounted cytospins of colchicine treated HL60 cells, where blocked metaphase 1 is short, thick chromosomes, and blocked metaphase 2 is fused chromosomes. Nota bene (N.B.) similar results were obtained with OCI-AML3 cells. |
Cells were classified as normal metaphases if the chromosomes line up along the equator. Blocked metaphases were distinguished by the presence of short, thick, and in some cases fused chromosomes, and the absence of a metaphase plate.
Since colchicine concentration and exposure time were the same for OCI-AML3 and HL60 treated cells. Thus the cell proliferative potential for each cell type was estimated by the percent of the blocked metaphases after 4 h exposure to colchicine. The percent of normal metaphases was used as an indicator for the proliferative potential of the untreated controls. To calculate the percentage of metaphases the slides were viewed with the immunofluorescence microscope and the total number of cells in a single field was counted. Then the same field was scanned again and only cells in mitosis were counted and categorized. To obtain sufficiently accurate values for metaphases percentage, metaphases were calculated in four different cytospins and their mean was mathematically estimated.
Intriguingly, we observed that the percentage of the blocked metaphases of the colchicine treated OCI-AML3 and HL60 cells (29.8% and 21.6%, respectively) was significantly higher than their equivalent untreated controls (3.1% and 8%, respectively), as shown in Figure 2. This denotes that the percentage of the blocked metaphases is a good indicator of the cell proliferative potential within 4 h period. However, the difference in the proliferative potential of the colchicine OCI-AML3 and the untreated control was significantly higher than that for the colchicine HL60.
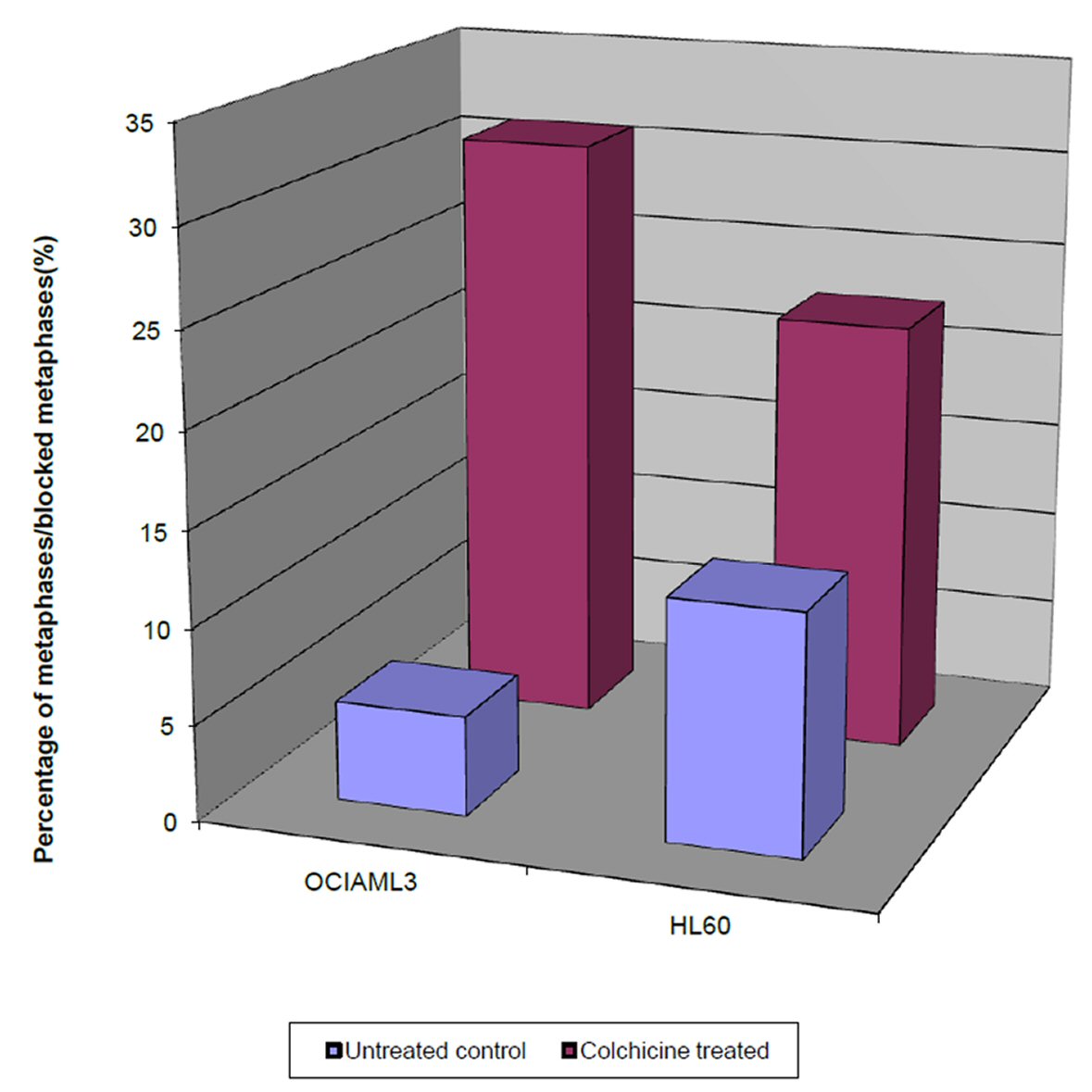 Click for large image | Figure 2. Single parameter histograms of the percentage of the blocked metaphases in colchicine treated OCI-AML3 and HL60 cells after 4 h, and the percentage of normal metaphases in untreated OCI-AML3 and HL60, after the same duration. |
On the other hand, the percentage of the blocked metaphases of the colchicine treated OCI-AML3 was significantly higher than that of the treated HL60. Taken together these results showed that the cell proliferative potential of the colchicine OCI-AML3 cells is significantly higher than that of colchicine HL60 cells.
Kinetic changes of NPM during proliferation of AML derived cells
Immunostaining with NPM FC 61991 showed that wtNPM is nucleolarly localized in resting cells and is translocated into the cytoplasm during cell division. This translocation was found to be cell cycle dependent, as wtNPM was detected in the nucleoli of interphase cells including G1, S, and G2 phases. Cytoplasmic translocation of wtNPM was detected in cells in active phases of cell division including prophase, metaphase, blocked metaphase, and anaphase cells (Fig. 3a, b). However, wtNPM was noted to relocalize gradually into the nucleoli during the telophase which represents the end of mitosis.
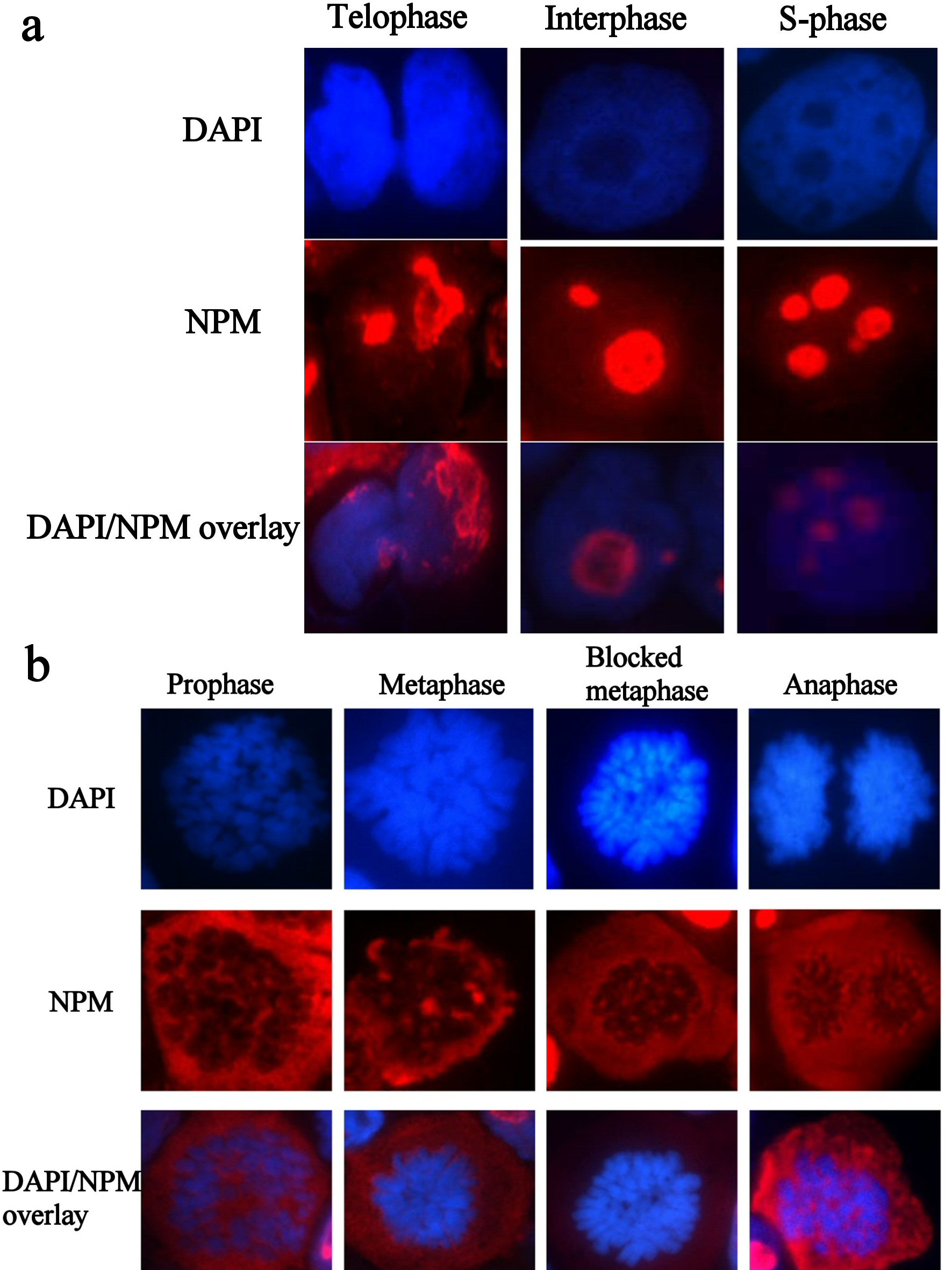 Click for large image | Figure 3. (a) Immunofluorescence images of resting HL60 cells, immunostained with wtNPM (FC61991), with TR (red) conjugated secondary antibody, and nuclei were stained with DAPI (blue). N.B. similar images were obtained with colchicine treated and control cytospins. (b) Immunofluorescence images of actively dividing HL60 cells, immunostained with wtNPM (FC61991), with TR (red) conjugated secondary antibody, and nuclei were stained with DAPI (blue). N.B. similar images were obtained with colchicine treated and control cytospins. |
Wild type NPM translocation during proliferation was found to be unrelated to the duration and extent of colchicine effect. Furthermore, these findings were similar among the untreated and colchicine treated HL60 and OCI-AML3 cultures.
In sharp contrast to wtNPM, immunostaining with ab15440 showed nucleolar and a nucleocytoplasmic localization in OCI-AML3 even during G0 phase (Fig. 4). This appearance was not seen in HL60 cells, and was not seen using antibodies that did not detect mNPM, strongly suggesting that the nucleoplasmic and cytoplasmic NPM was of mNPM type. However, the cytoplasmic localization was detected in actively dividing cells of both types. Comparing these findings with the results of wt immunostaining and the untreated controls denote that the cytoplasmic staining in the dividing HL60 represents the cytoplasmic translocation of wtNPM that was discussed before.
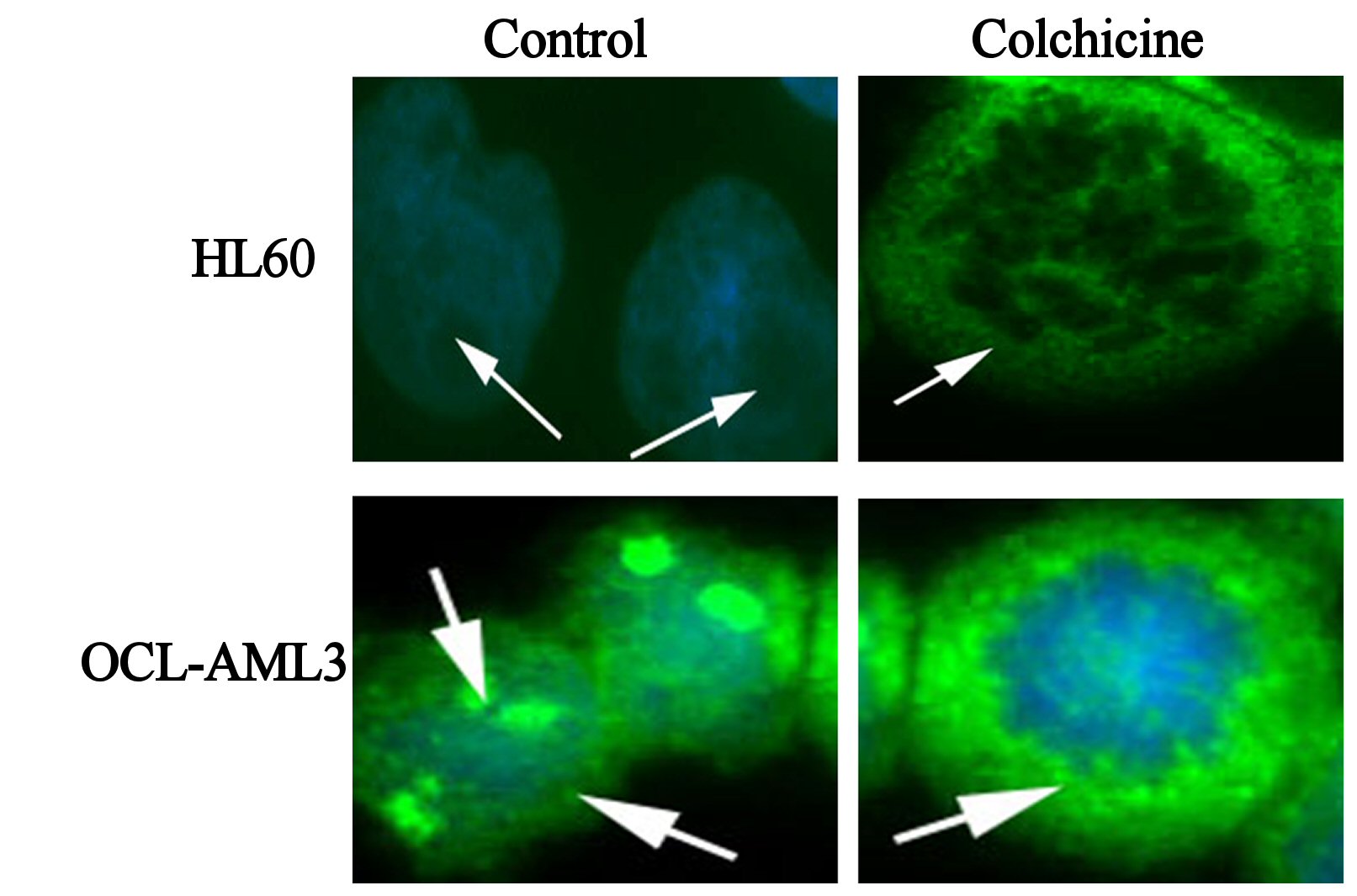 Click for large image | Figure 4. Immunofluorescence images of control, colchicine treated HL60 and OCI-AML3, as indicated in the figure, immunostained with antiNPM (ab 15440) with FITC (green) conjugated secondary antibody, and nuclei were stained with DAPI (blue). |
Nucleophosmin expression during proliferation of AML derived cells
To determine NPM expression during AML proliferation, proteomic analysis of NPM was conducted in WCL of colchicine treated and untreated HL60 and OCI-AML3. The identity of NPM was confirmed by detection of a 37-kD band, after immunoblotting with specific NPM antibody.
Immunoblotting of anti-NPM FC61991 in lysates of HL60 and OCI-AML3, both colchicine treated and untreated showed barely difference in wtNPM expression between HL60 and OCI-AML3. Moreover, the cytoplasmic translocation of wtNPM during cellular proliferation was not accompanied with changes in its protein levels and wtNPM protein expression in G0 cells was similar to that in dividing cells of both cell types as depicted in Figure 5.
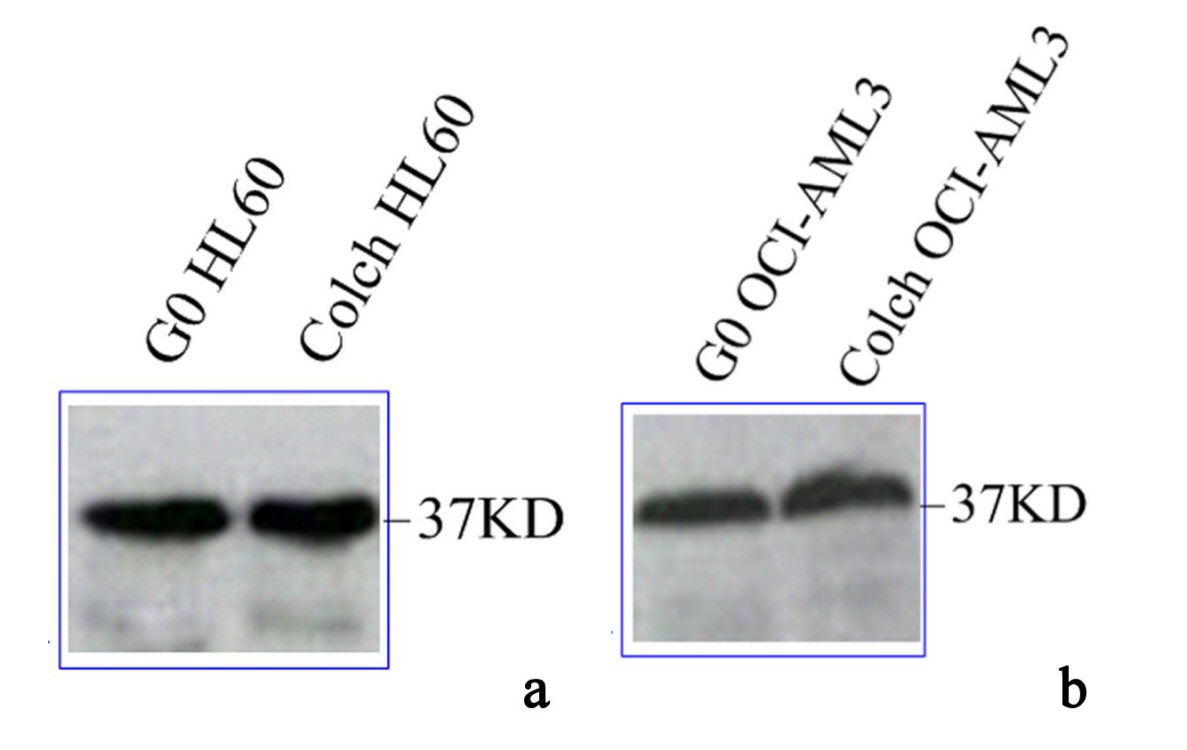 Click for large image | Figure 5. Immunoblotting results of wtNPM in lysates of (a) G0 and colch HL60, (b) G0 and colch OCI-AML3, results were scanned and edited with Epson scanner and program. |
Results of western blotting immunoassay of anti-NPM antibodies (NA24 and ab1544) in protein extracts of OCI-AML3 and HL60, both colchicine treated and untreated are shown in Figure 6. These results were inconsistent with our immunofluorescence findings, as there was no significant difference between total NPM protein expression between OCI-AML3 and HL60. The most interesting finding was a significant reduction in total NPM protein in colchicine treated lysates compared with the untreated resting ones, this finding was the same in both cell types.
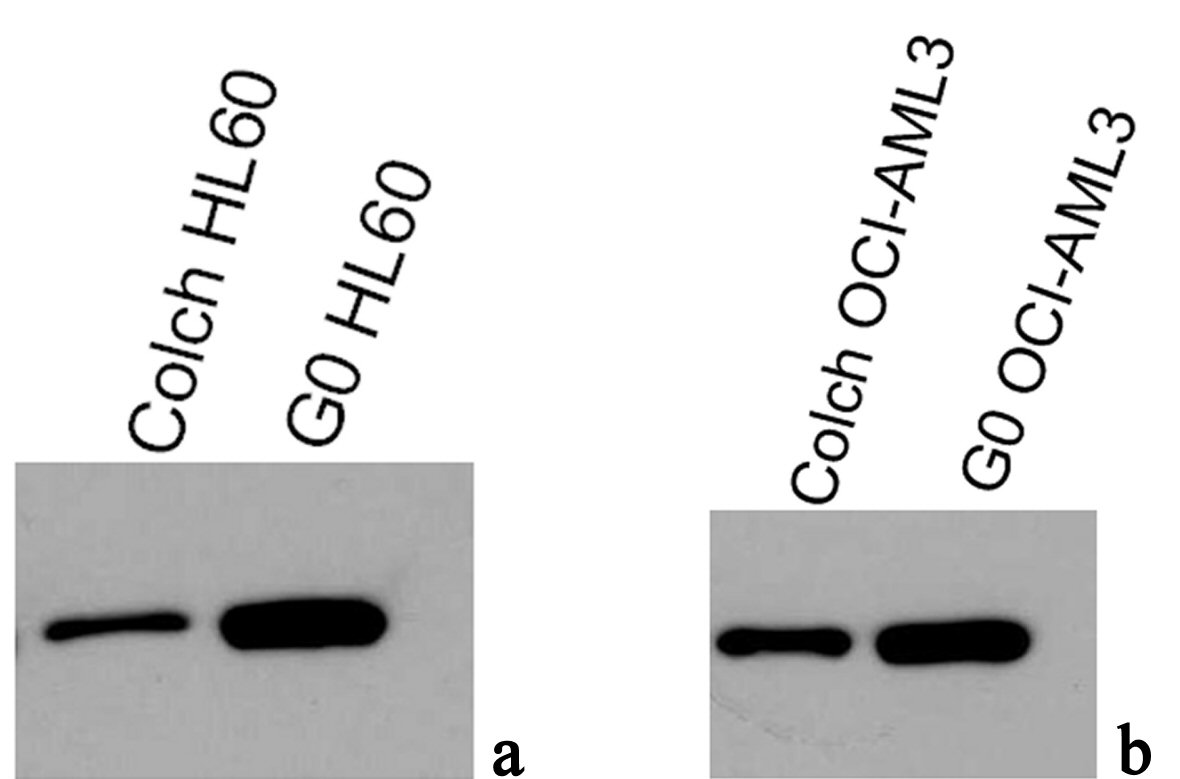 Click for large image | Figure 6. Immunoblotting results of antiNPM (ab15440) in lysates of (a) G0 and colch HL60, (b) G0 and colch OCI-AML3, results were scanned and edited with Epson scanner and program. |
| Discussion | ▴Top |
Although NPM gene mutation has been implicated by others in AML pathogenesis, how mNPM contributes in AML development remains unclear [10]. In the present study, a thorough investigation of NPM during proliferation of AML derived cells was done. The main aim is to have a complete scenario of the contribution of NPM gene mutation in AML leukemogenesis. The study was conducted through an integration of viability studies, immunocytochemical and immunoblotting techniques.
The viability study results showed higher proliferative potential of cells expressing mNPM compared to those bearing wtNPM. This was noted in the colchicine treated cultures, while the proliferative potential of the normally growing cultures was higher in those bearing wtNPM. This denoted the presence of other differences besides mNPM expression that could explain these results. One of these is the difference in the cell cycle kinetics between the two types of cells.
These findings are supported by the findings of others who found that mNPM promotes proliferation of myeloid cells in zebra fish embryo and in mice [11, 12].
How mNPM enhanced the proliferation of OCI-AML3 cells could be explained by the previous findings of the role of NPM in cellular proliferation. NPM was found to play critical roles in the control of different aspects of cell growth and homeostasis, such as ribosome biogenesis, centrosome duplication, and cell cycle progression [10]. Moreover, the cytoplasmically localized mNPM was proposed to downregulate the ARF tumor suppressor gene and enhancing the c-MYC oncogene [13-15].
The immunofluorescence and immunoblotting studies showed that wtNPM is localized in the central granular region of the nucleolus in resting cells. Then it gradually translocated to the cytoplasm in active mitotic cells and by the end of mitosis relocalized to the nucleoli. However, these kinetic changes were not accompanied with changes in NPM protein levels. On the other hand mNPM showed no changes either in its localization or in its expression, during cellular proliferation.
Our findings regarding the cytoplasmic translocation of wtNPM are consistent with the findings of others [16, 17]. However, the present study demonstrated how this translocation is different from one cell cycle phase to another. It also showed no or little effect of the cell cycle phases on mNPM localization. NPM cytoplasmic translocation was considered to play direct or indirect role on AML pathogenesis, either through disturbing NPM functions or binding with other cytoplasmic proteins [18, 19].
In conclusion, this study demonstrated that mNPM offered a proliferative advantage to cells bearing the mutation, and contributed in AML leukemogenesis and development. However, further studies are still needed to investigate the role of mNPM in proliferation of freshly prepared primary AML cells.
Acknowledgments
Thanks to Dr. Karen-Rees Unwin at the Manchester School of Medicine, UK, and to Dr. Suzanne Johnson, at Manchester School of Medicine UK, for their valuable guidance throughout the work. Great thanks to the Egyptian Cultural Bureau in London for their financial and other types of support to the author throughout the research work. Thanks to teacher Jennifer Huerta Mesde at Shaqra University, KSA for her efforts in revision of English language editing.
Grant Support
This work was supported by the Egyptian Cultural Bureau Educational Grant.
Conflict of Interest
There was no conflict of interest associated with this work.
| References | ▴Top |
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342-348.
doi pubmed - Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264-278.
doi pubmed - Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254-266.
doi pubmed - Arber DA, Brunning RD, Le Beau MM, et al. Acute myeloid leukaemia with recurrent genetic abnormalities. In: Swerdlow SH, Campo E, Harris NL, et al, eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2008:110-123.
- Mattsson G, Turner SH, Cordell J, Ferguson DJ, Schuh A, Grimwade LF, Bench AJ, et al. Can cytoplasmic nucleophosmin be detected by immunocytochemical staining of cell smears in acute myeloid leukemia? Haematologica. 2010;95(4):670-673.
doi pubmed - Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes Cancer. 2010;1(7):681-689.
doi pubmed - Alegria-Schaffer A. Western blotting using chemiluminescent substrates. Methods Enzymol. 2014;541:251-259.
- Quentmeier H, Martelli MP, Dirks WG, Bolli N, Liso A, Macleod RA, Nicoletti I, et al. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia. 2005;19(10):1760-1767.
doi pubmed - Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350-4354.
doi pubmed - Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493-505.
doi pubmed - Cheng K, Sportoletti P, Ito K, Clohessy JG, Teruya-Feldstein J, Kutok JL, Pandolfi PP. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood. 2010;115(16):3341-3345.
doi pubmed - Bolli N, Payne EM, Grabher C, Lee JS, Johnston AB, Falini B, Kanki JP, et al. Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood. 2010;115(16):3329-3340.
doi pubmed - Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, Volorio S, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66(6):3044-3050.
doi pubmed - den Besten W, Kuo ML, Williams RT, Sherr CJ. Myeloid leukemia-associated nucleophosmin mutants perturb p53-dependent and independent activities of the Arf tumor suppressor protein. Cell Cycle. 2005;4(11):1593-1598.
doi pubmed - Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol. 2008;182(1):19-26.
doi pubmed - Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, Nicoletti I. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23(10):1731-1743.
doi pubmed - Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56(3):379-390.
doi - Bolli N, De Marco MF, Martelli MP, Bigerna B, Pucciarini A, Rossi R, Mannucci R, et al. A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF. Leukemia. 2009;23(3):501-509.
doi pubmed - Liso A, Bogliolo A, Freschi V, Martelli MP, Pileri SA, Santodirocco M, Bolli N, et al. In human genome, generation of a nuclear export signal through duplication appears unique to nucleophosmin (NPM1) mutations and is restricted to AML. Leukemia. 2008;22(6):1285-1289.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


