| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website http://www.thejh.org |
Short Communication
Volume 7, Number 4, December 2018, pages 149-153
Functional Properties of Circulating Exosomes Mediated by Surface-Attached Plasma Proteins
Tatiana Shtama, b, c, Stanislav Naryzhnyc, d, Arthur Kopylovd, Elena Petrenkod, Roman Samsonova, b, Roman Kamyshinskye, Yana Zabrodskayac, Daniil Nikitinf, Maxim Sorokine, g, h, Anton Buzdinf, g, h, i, Anastasia Maleka, b, j
aN.N.Petrov National Medical Research Center of Oncology, 197758, Leningradskaya 68, St.-Petersburg, Russia
bLtd Oncosystem, 143026, Lugovaya 4, Skolkovo Innovation Center, Moscow, Russia
cPetersburg Nuclear Physics Institute named by B.P. Konstantinov of National Research Centre «Kurchatov Institute», 188300, Orlova roscha 1, Gatchina, Russia
dOrekhovich Institute of Biomedical Chemistry of Russian Academy of Medical Sciences, 119121, Pogodinskaya 10, Moscow, Russia
eNational Research Center “Kurchatov Institute”, 123098, Academician Kurchatov Square 1, Moscow, Russia
fEngelhardt Institute of Molecular Biology, Russian Academy of Sciences, 119991, 32, Vavilova Str., Moscow, Russia
gShemyakin-Ovchinnikov Institute of Bioorganic Chemistry, 117997, 16/10 Miklukho-Maklaya Str., Moscow, Russia
hOmicsWay Corp., 91789, 340 S Lemon Ave, Walnut, CA, USA
iI.M. Sechenov First Moscow State Medical University (Sechenov University), 119991, 8-2 Trubetskaya St., Moscow, Russia
jCorresponding Author: Anastasia Malek, Oncoendocrinology Laboratory, N.N.Petrov National Medical Research Center of Oncology, 197758, Leningradskaya 68, St.-Petersburg, Russia
Manuscript submitted May 1, 2018, accepted October 8, 2018
Short title: Plasma Exosome Mediated by Surface Proteins
doi: https://doi.org/10.14740/jh412w
| Abstract | ▴Top |
Background: Exosomes and other types of extracellular vesicles present an important component of circulating plasma. Exosomes released by endothelial and blood cells account for majority of plasma exosomal population; exosomes secreted by other cells might cross tissue-plasma barrier and reach circulating plasma as well. Definitely, exosomes of different cellular origins are different by content and function. However, exosomal surface membrane interacts with plasma components. This interaction may alter composition of exosomal surface and hence, provide these vesicles with new functional properties. This study was aimed to estimate composition and possible functional role of proteins attached on the surface of plasma exosomes.
Methods: Here, extracellular vesicles from human plasma were isolated by ultracentrifugation and treated by trypsin. Trypsinized and native exosomes were analyzed by nanoparticle tracking analysis, Western blotting and quantitative high-resolution mass spectrometry.
Results: Surface-attached proteins were removed from exosomes isolated from plasma of healthy donors by incubation with serine protease (trypsin). Treatment did not impact exosomes integrity while slightly reduced hydrodynamic radius. Mass spectrometry revealed 259 exosomal proteins; among them 79 proteins were completely removed and more than half of the proteins were partially removed by trypsinization. Gene ontology functional annotation revealed mostly extracellular locations of proteins cleaved from a surface of the plasma exosomes. Moreover, proteins cleaved from the exosome surface are supposed to be implicated into integrin-linked kinase (ILK), focal adhesion kinase (FAK) and other pathways connecting cell surface with intracellular signaling cascades.
Conclusion: Taken together, our results demonstrate that a surface of circulating exosomes is decorated by plasma proteins, and these proteins can mask tissue-specific characteristic of the exosomal surface membrane and provide exosomes with new and uniform properties.
Keywords: Exosomes; Plasma proteins; Mass spectrometry; FAK signaling; ILK signaling
| Introduction | ▴Top |
Exosomes are a sort of extracellular vesicles (EVs) derived from endosomal system and released out of cells upon fusion of multi-vesicular endosomes with a cell membrane. Exosomes differ from other types of EVs by relatively small size (50 - 130 nm) and expression of specific exosomal markers (CD9, CD63 and others). It was assumed earlier that exosomes involved in clearance of cellular waste, while recently exosomes are attracting considerable attention as mediators of intercellular communication [1]. Exosomes can be found in most of biological fluids. Concentration of the plasma exosomes isolated by differential centrifugation and measured by nanoparticle tracking analysis (NTA) is 9.5 × 1010 ± 1 × 1010 vesicles/mL [2] that is equivalent to about 0.1-0.3% of the plasma proteins (166 ± 50 µg/mL). Assuming high level and variable aspects of functional activity, plasma exosomes might represent an important component of the blood in terms of its defensive, homeostatic and signal transduction properties.
The plasma exosomes have been investigated by different approaches including mass spectrometry [3]. Proteomics of plasma exosomes can reveal specific subsets of proteins reflecting various cellular functions. For instance, protein profile of plasma vesicles resembled protein profiles of vesicles from platelets, antigen-presenting cells, and natural killer T cells [4]. Moreover, cancerous disease or other systemic pathologies may affect composition of blood-cell derived vesicles in plasma or even change total content of plasma exosomes [4]. Currently, plasma circulating exosomes are considered as a promising source of markers for diagnostic and monitoring of various illnesses including cancer [5]. Despite great effort done in exosomes research, there are still considerable challenges associated with heterogeneity of plasma exosomes including cellular origin, molecular composition and functions [6]. Individual variability of donors [7] and methodological preferences of researchers [8] also introduce certain bias into exosome research.
One possible approach to identify different populations of plasma exosomes is subtraction of common characteristics. For instance, separation of exosomal samples from contaminating plasma lipoproteins allowed detailed analysis of exosomal proteome and discovery of new exosome-associated proteins [9]. Besides lipoproteins, other plasma components, like signal peptides or proteins, are interacting with circulating exosomes. Thus, plasma proteins can attach to the surface of vesicles and provide them with certain uniformity. This assumption is in line with results of recent study: serum exosome proteome is represented by extracellular proteins by 23% [10]. Extracellular components absorbed to the vesicular membrane during circulation can mask initial diversity of the exosomes and even provide them with universal functional properties. For example, fibronectin (FN) attached to exosomes is shown to be involved in various aspects of exosomal function [11, 12]. This study was aimed to estimate composition and possible functional role of proteins attached to the surface of plasma exosomes.
| Material and Methods | ▴Top |
Plasma collection, isolation and characterization of EVs
Design of the study and use of human material was approved by local research ethics committee of N.N. Petrov National Medical Research Center of Oncology (no. 4-14/13.02.2014). All participants have signed informed consent form before collection of the biological material (blood). In order to avoid possible impact of gender difference in content and quantity of plasma vesicles, only female donors were included in the study. Blood samples were collected from 10 healthy donors in the department of blood transfusion of N.N. Petrov National Medical Research Center of Oncology using EDTA-coated vacutainers. Exosomes were isolated by differential centrifugation, as previously described [13]. The size, homogeneity and concentration of isolated exosomes were evaluated by dynamic light scattering (DLS) and NTA according to the manufacturer’s instructions (Malvern, UK). Techniques of atomic force microscopy (AFM) and cryo-electron microscopy (cryo-EM) were used for the direct visualization of exosomal particles, as described earlier [14, 15]. The presence of exosomal markers (CD63, CD9) was assayed by flow cytometry with Exo-FACS ready-to-use kit for human exosome analysis (HansaBioMed, Estonia).
Enzymatic modification and analysis of plasma exosomes
Exosomes isolated from plasma by ultracentrifugation were treated with 0.25% trypsin for 1 h, washed with phosphate-buffered saline (PBS) and purified again. Trypsinized and intact exosomes were analyzed by NTA, Western blotting (WB) and mass spectrometry (MS). For WB, protein concentration in extracts was normalized using Bradford method (Bio-Rad, USA). WB was performed as described [13] with primary antibodies against FN (ab2413; Abcam, USA) and CD63 (ab68418; Abcam, USA) followed by a peroxidase-labeled secondary antibody (A0545; Sigma, Germany). MS (shotgun analysis) of exosomal proteins was performed according to the filter-aided sample preparation (FASP) protocol [16, 17]. Briefly, the proteins were digested with trypsin (Promega Trypsin Gold), and MS/MS analysis of resulting peptides was performed in duplicate using an Orbitrap Fusion Lumos MS (Thermo Scientific, USA) [18, 19].The data were searched by Mascot 2.4.1 (http://www.matrixscience.com/). As a protein sequence database, NeXtProt (October 2014) was used. To be included in the list of exosomal proteins, we considered only the proteins with high-quality identifications (≥ 2 peptides, 95 % confidence at least in one sample (treated or not treated by tripsin)). The exponentially modified form of protein abundance index (emPAI) defined as the number of identified peptides divided by the number of theoretically observable tryptic peptides for each protein was used to estimate protein abundance [20, 21].
Gene ontology (GO) functional annotation and pathway analysis
GO analysis of exosomal proteins was performed with functional annotation tool of The Database for Annotation, Visualization and Integrated Discovery (DAVID) [22, 23]. Human molecular pathways were extracted from the following databases: BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), KEGG [24], NCI [25], Reactome [26] and SABiosciences Pathway Central (http://www.sabiosciences.com/pathwaycentral.php). We calculated pathway activation score (PAS) for each pathway from the above-mentioned databases. PAS of the pathway P is the number of exosome surface genes, which are involved in the pathway P. P value calculation was performed using DAVID software (Pubmed PMID: 19131956). Briefly, P value was calculated with modified Fisher’s exact test (column “P value”, Supplementary Table 1). Adjustment for multiple testing was performed using the Benjamini-Hochberg procedure (column “False Discovery Rate corrected P-value”, Supplementary Table 1).
| Results and Discussion | ▴Top |
Surface of circulating exosomes is decorated by various extracellular proteins
Isolated vesicles were characterized in accordance with International Society for Extracellular Vesicles (ISEV) guidelines [27]. Size of vesicles was in a range of 70 - 200 nm, as it was measured by DLS (Fig. 1a). The morphology of single vesicles was assayed by AFM and cryo-EM (Fig. 1c, d). Presence of exosomal markers (CD9, CD63) on the surface of vesicles was confirmed by flow cytometry (Fig. 1b). Thus, nanovesicles isolated from human plasma had a typical spherical form, contained a bilayer membrane, were enriched with exosomal markers and were considered to be exosomes. Freshly isolated and characterized exosomes from 10 individuals were pooled in equivalent quantities. Afterwards, half amount of pooled samples was treated with 0.25% trypsin solution and half amount was processed without trypsinization. Thus, treatment with trypsin led to reduction of vesicles size (Fig. 2a). Moreover, by NTA it is possible to estimate the intensity of eradiation reflected from the surface of the analyzed particles. As it can be seen by comparing the top and bottom panel (Fig. 2b), trypsinization leads to significant change in the structure of the exosome surface. It is logical to assume that such a treatment with trypsin leads to the complete or partial removal of proteins adhered to the surface of vesicles. For example, FN is a high abundance protein of plasma and can non-specifically bind to the surface of the circulating vesicles, regardless of their origin. To test this hypothesis, equivalent number of intact and treated with trypsin particles was used for the analysis of exosomal marker CD63 and FN by WB (Fig. 2c). As expected, incubation with trypsin leads to the destruction of exosomal FN, but did not significantly affect the content of transmembrane tetrasponine CD63. Next, we aimed to evaluate the whole spectrum of proteins attached to the surface of circulating exosomes by mass spectrometry. We reported here more than 250 high quality (≥ 2 peptides, 95% confidence) identifications of human exosomal proteins (Supplemenatry Table 2). Abundancy (emPAI) of more than half of these proteins was being reproducibly reduced after processing with trypsin. Besides, 79 proteins were completely removed by trypsinization. (Supplementary Table 2). FN isoform 1 (FN1) is plasma abundant protein that was identified by MS in intact plasma exosomes and was almost completely cleaned from exosomes by trypsin. These results were confirmed by WB (Fig. 2c).
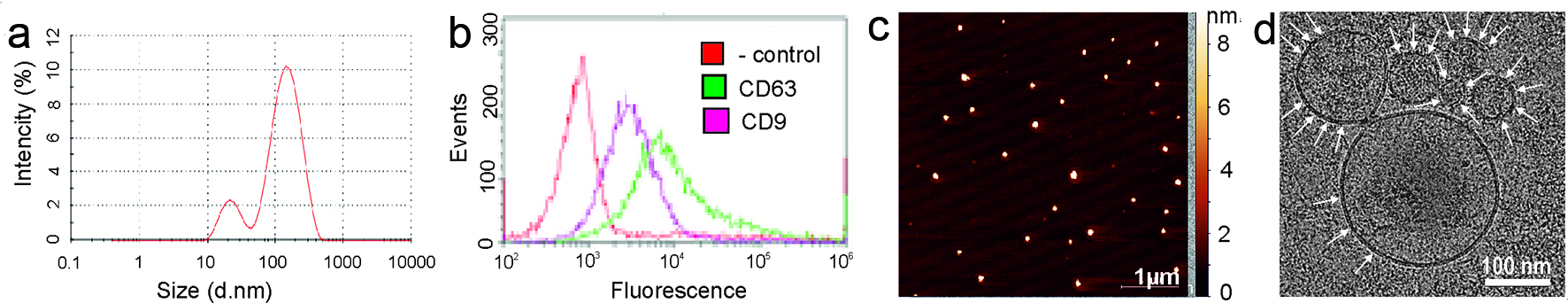 Click for large image | Figure 1. Exosomes characteristics. (a) Exosome size was estimated by dynamic light scattering (DLS). (b) Flow cytometry analysis of exosomes for the surface expression of exosomal markers CD9 and CD63 was carried out by ready to use kit for FACS analysis of purified exosomes (HansaBioMed). Negative control Exo(-) was performed without any vesicles. (c) The surface topography of plasma exosomes obtained with atomic force microscopy (AFM). The scale bar is 1 µm. On the right is the pseudocolor ruler indicating the particles’ height (nm). (d) Cryo-electron microscopy (Cryo-EM) of the exosomes derived from plasma. The scale bar is 100 nm. |
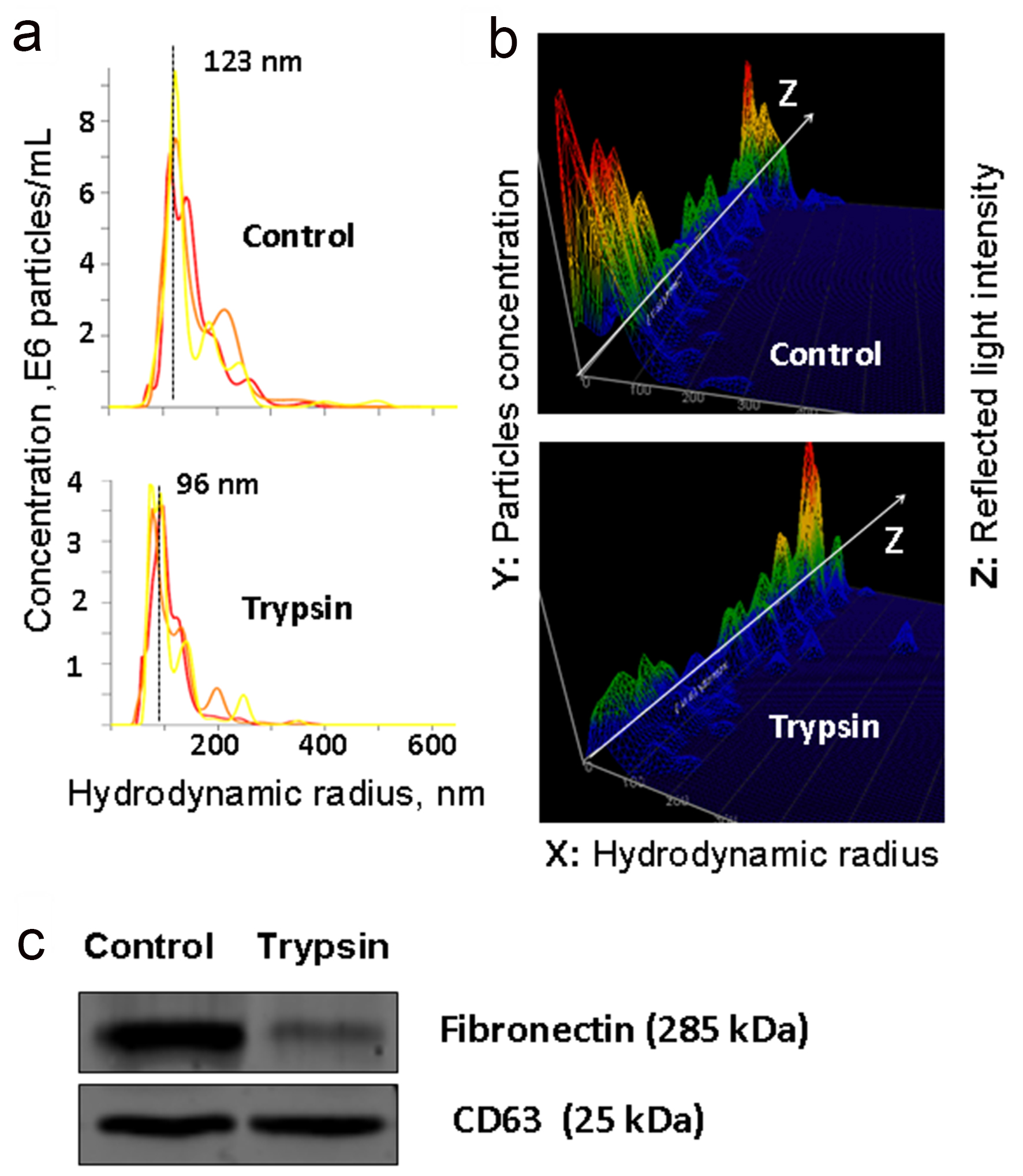 Click for large image | Figure 2. Analysis of trypsinized and intact plasma exosomes. (a and b) The results of nanoparticle tracking analysis (NTA) of exosomes isolated from plasma and treated with trypsin: the ratio of the size and concentration of exosomes (a); a three-dimensional graph, the Z axis shows changes in characteristics of light reflected from the surface of exosomes (b). (c) WB detection of fibronectin (FN) and exosomal marker CD63 in exosomes isolated from native plasma and exosomes treated with trypsin. |
Plasma proteins attached to exosomes surface are implicated in ILK- and FAK-signaling pathways
GO analysis revealed several GO terms significantly over-represented in a set of the proteins cleared from the exosomal surface. Thus, clusters of secreted extracellular proteins and proteins modified by disulfide bond or glycosylation were significantly enriched among exosome-attached proteins, with enrichment score of 12.29 (Supplementary Table 1). Other clusters were enriched less significantly (enrichment score: 5.66) with GO terms relevant to various aspect of cellular and humoral immunity as well as proteolytic plasma activity (Supplementary Table 1). Thus, exosomal surface is decorated by extracellular proteins that are abundantly present in plasma and are likely being attached to the exosomal membrane during circulation. It is not yet clear if interaction of plasma proteins with exosomes is selective in terms of exosome origin or chemical composition or all circulating vesicles are equally covered by plasma components. In any case, plasma proteins attached to an exosome surface may be involved in exosome-mediated cellular communication or can even provide exosomes with new regulatory functions. To predict intracellular pathways that may be affected by plasma proteins attached onto surface of circulating vesicles, pathway analysis was applied. List of intracellular regulatory cascades containing proteins cleared from a surface of the exosomes is presented in Supplementary Table 3. Signal transduction pathways mediated by integrin-linked kinase (ILK) and focal adhesion kinase 1 (FAK1) are predicted to be mostly affected by plasma exosomes. For examples, exosome-associated FN1, extracellular matrix protein 1 (ECM1) and filamin A (FLNA) can directly interact with heterodimeric transmembrane receptor integrin that is a vital cell adhesion receptor and an upstream activator of ILK-signaling cascade. Other five exosome-associated molecules can interfere with downstream events of ILK signaling (Fig. 3). Identified intracellular signaling pathways are involved in control of many various cellular functions including cell migration, adhesion [28], differentiation [29] (Fig. 3) and are essential for cell-to-cell communication and cell-to-extracellular matrix integration. Thus, our results highlight new aspect of regulatory function of plasma exosomes and need to be deeper investigated. It’s important to note that plasma proteins attached to the exosome surface may overcome new regulatory qualities as well as provide exosomes with new functions. Considering general character of observed phenomenon, it may have great impact into physiology and regulatory function of the blood.
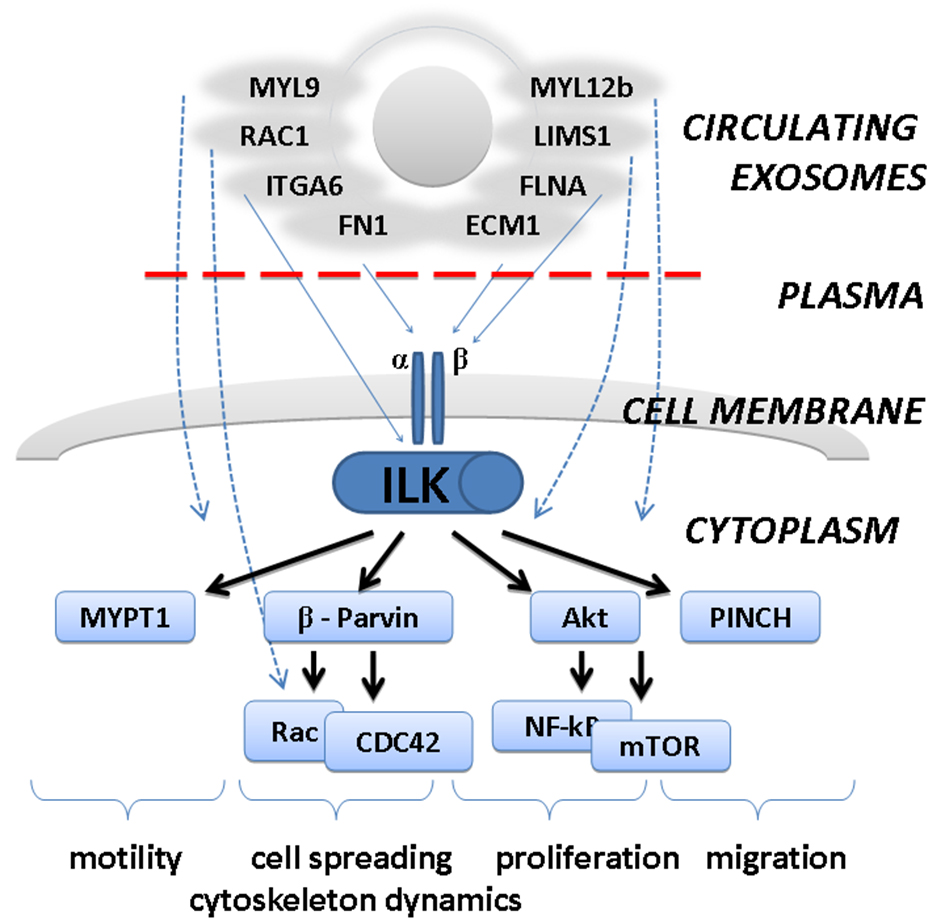 Click for large image | Figure 3. Predicted involvement of exosome-attached extracellular proteins in integrin-linked kinase (ILK) pathway. Plasma proteins attached to the surface of circulating exosomes may affect biology of the cells by interaction with surface receptors (integrins) and activation of downstream ILK-pathway. This would lead to stimulation of cellular migration, motility, proliferation, etc. |
Conflict of Interest
The authors declare no conflict of interest.
Grant Support
The study has been supported by RFBR (grants number 15-54-12380 and 18-015-00289). Cryo-EM experiments were supported by RSF (grant number 17-14-01416). Mass-spectrometry measurements were performed using the equipment of the Human Proteome Core Facilities of the Institute of Biomedical Chemistry (Russia), which is supported by the Ministry of Education and Science of the Russian Federation (agreement 14.621.21.0017, unique project ID RFMEFI62117X0017).
| References | ▴Top |
- Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226-1232.
doi pubmed - Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. 2014;411:55-65.
doi pubmed - Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267-283.
doi pubmed - Koliha N, Heider U, Ozimkowski T, Wiemann M, Bosio A, Wild S. Melanoma affects the composition of blood cell-derived extracellular vesicles. Front Immunol. 2016;7:282.
doi pubmed - Whiteside TL. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev Mol Diagn. 2015;15(10):1293-1310.
doi pubmed - Tkach M, Kowal J, Thery C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos Trans R Soc Lond B Biol Sci. 2018;373(1737):20160479.
- Bastos-Amador P, Royo F, Gonzalez E, Conde-Vancells J, Palomo-Diez L, Borras FE, Falcon-Perez JM. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J Proteomics. 2012;75(12):3574-3584.
doi pubmed - Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13(22):3354-3364.
doi pubmed - Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, Lotvall J, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75(15):2873-2886.
doi pubmed - Koller A, Patel P, Kim JK, Chen EI. Proteomics analysis of circulating serum exosomes. Methods Mol Biol. 2017;1619:213-225.
doi pubmed - Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem. 2016;291(4):1652-1663.
doi pubmed - Dismuke WM, Klingeborn M, Stamer WD. Mechanism of fibronectin binding to human trabecular meshwork exosomes and its modulation by dexamethasone. PLoS One. 2016;11(10):e0165326.
doi pubmed - Samsonov R, Burdakov V, Shtam T, Radzhabova Z, Vasilyev D, Tsyrlina E, Titov S, et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. 2016;37(9):12011-12021.
doi pubmed - Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov MV. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88.
doi pubmed - Shtam TA, Samsonov RA, Volnitskiy AV, Kamyshinsky RA, Verlov NA, Kniazeva MS, Korobkina EA, et al. [Isolation of extracellular micro-vesicles from cell culture medium: comparative evaluation of methods]. Biomed Khim. 2018;64(1):23-30.
doi pubmed - Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359-362.
doi pubmed - Naryzhny S, Maynskova M, Zgoda V, Archakov A. Dataset of protein species from human liver. Data Brief. 2017;12:584-588.
doi pubmed - Naryzhny S, Zgoda V, Kopylov A, Petrenko E, Kleist O, Archakov A. Variety and dynamics of proteoforms in the human proteome: aspects of markers for hepatocellular carcinoma. Proteomes. 2017;5(4):33.
doi pubmed - Naryzhny S, Maynskova M, Zgoda V, Archakov А. Zipf's law in proteomics. J Proteomics Bioinform. 2017;10:79-84.
doi - Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265-1272.
doi pubmed - Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102.
doi pubmed - Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57.
doi pubmed - Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1-13.
doi pubmed - Nakaya A, Katayama T, Itoh M, Hiranuka K, Kawashima S, Moriya Y, Okuda S, et al. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41(Database issue):D353-357.
pubmed - Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674-679.
doi pubmed - Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472-477.
doi pubmed - Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913.
doi pubmed - Pan L, Zhao Y, Yuan Z, Qin G. Research advances on structure and biological functions of integrins. Springerplus. 2016;5(1):1094.
doi pubmed - Vitillo L, Kimber SJ. Integrin and FAK regulation of human pluripotent stem cells. Curr Stem Cell Rep. 2017;3(4):358-365.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


